Insights into the molecular mechanisms of browning tolerance in luffa: a transcriptome and metabolome analysis
- PMID: 40556689
- PMCID: PMC12186849
- DOI: 10.3389/fpls.2025.1530531
Insights into the molecular mechanisms of browning tolerance in luffa: a transcriptome and metabolome analysis
Abstract
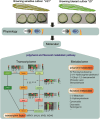
Introduction: Enzymatic browning significantly affects the edible, nutritional, and commercial value of luffa. Investigating the expression and regulation of key enzyme genes involved in the browning process is crucial for understanding the molecular mechanisms underlying luffa browning.
Methods: Fruit samples were collected at 15 (S1), 20 (S2), and 45 days (S3) after flowering from two contrasting luffa varieties: the browning-sensitive Long-quan-yi (LQY) and the browning-tolerant Jiang-du (JD). RNA-sequencing technology, combined with ultra-performance liquid chromatography electrospray ionization tandem mass spectrometry (UPLC-ESI-MS/MS), was used to obtain transcriptome and metabolome data, which were subsequently analyzed using a series of bioinformatics approaches. Quantitative polymerase chain reaction (q-PCR) was used to validate gene expression.
Results: Compared with JD, the ROS levels and PPO activity were elevated in LQY. In the polyphenol metabolic pathway, 24 key enzyme genes including CuAO, PPO, and TDC, were identified. In the flavonoid metabolic pathway, 57 key structural genes, such as PAL, C3H, and 4CL, were identified. These genes showed different expression patterns between the two luffa varieties. Differentially expressed genes were mainly involved in the regulation of 34 MYB, 15 bHLH, 19 WD40, and 14 WRKY transcription factors. Further metabolomics analysis showed that the levels of polyphenol metabolites were upregulated in LQY, whereas the levels of flavonoid metabolites were upregulated in JD.
Discussion: This study integrated transcriptomic and metabolomics data to identify key genes, transcription factors and metabolic pathways associated with luffa browning. q-PCR analysis was performed to validate the expression of POD and MYB genes. These findings provide a theoretical foundation for further investigation into the molecular mechanisms underlying luffa browning and offer potential targets for genetic improvement or breeding strategies to enhance luffa quality.
Keywords: antioxidant enzyme; browning; flavonoid metabolism; metabolome; transcription factor; transcriptome.
Copyright © 2025 Wu, Sun and Lian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Understanding mechanisms of Polygonatum sibiricum-derived exosome-like nanoparticles against breast cancer through an integrated metabolomics and network pharmacology analysis.Front Chem. 2025 Jun 6;13:1559758. doi: 10.3389/fchem.2025.1559758. eCollection 2025. Front Chem. 2025. PMID: 40547857 Free PMC article.
-
Deciphering Shared Gene Signatures and Immune Infiltration Characteristics Between Gestational Diabetes Mellitus and Preeclampsia by Integrated Bioinformatics Analysis and Machine Learning.Reprod Sci. 2025 Jun;32(6):1886-1904. doi: 10.1007/s43032-025-01847-1. Epub 2025 May 15. Reprod Sci. 2025. PMID: 40374866
-
Metabolome and transcriptome profiling reveals light-induced anthocyanin biosynthesis and anthocyanin-related key transcription factors in Yam (Dioscorea Alata L.).BMC Plant Biol. 2025 May 30;25(1):729. doi: 10.1186/s12870-025-06738-w. BMC Plant Biol. 2025. PMID: 40442608 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
References
-
- Bustos M. C., Mazzobre M. F., Buera M. D. P. (2015). Stabilization of refrigerated avocado pulp: Effect of Allium and Brassica extracts on enzymatic browning. LWT-Food Sci. Technol. 61, 89–97. doi: 10.1016/j.lwt.2014.11.026 - DOI
LinkOut - more resources
Full Text Sources

