Exploring the complex interplay between oral Fusobacterium nucleatum infection, periodontitis, and robust microRNA induction, including multiple known oncogenic miRNAs
- PMID: 40558049
- PMCID: PMC12282186
- DOI: 10.1128/msystems.01732-24
Exploring the complex interplay between oral Fusobacterium nucleatum infection, periodontitis, and robust microRNA induction, including multiple known oncogenic miRNAs
Abstract
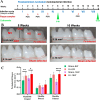
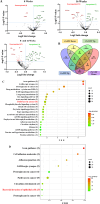
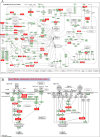
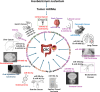
Fusobacterium nucleatum (F. nucleatum) is an oral commensal bacterium that can become pathogenic and is associated with periodontitis, adverse pregnancy outcomes, and colorectal cancer (CRC). MicroRNAs are conserved, non-coding RNA molecules that regulate gene expression and are detected in microbial infections. This study aims to characterize the microRNA expression kinetics in the mandibles of C57BL/6J mice infected with F. nucleatum for 8 and 16 weeks and to identify miRNAs as potential biomarkers using NanoString nCounter miRNA panels. Mice were divided into four groups: 8 weeks of infection, 16 weeks of infection, and their respective sham infection. F. nucleatum-infected mice showed 100% bacterial colonization on the gingival surface, along with a significant increase in alveolar bone resorption (P < 0.0001) and intravascular dissemination to the heart, indicating its invasive potential. Out of 577 miRNAs analyzed, seven miRNAs were upregulated, and two miRNAs were downregulated in the 8 weeks of infection group. In the 16 weeks of infection group, seven miRNAs were upregulated while 13 miRNAs were downregulated. Notably, miR-205, miR-210, and miR-199a-3p were differentially expressed at 8 weeks as well as miR-28 at 16 weeks and have been previously reported in human periodontitis. The 13 miRNAs induced by F. nucleatum (e.g., miR-361-5p) are linked to 13 multiple malignancies. In addition, miR-126-5p has been identified as a potential biomarker for patients with PD and cardiovascular disease. These results indicate that F. nucleatum induces several PD-related miRNAs and links them to systemic comorbidities. Furthermore, this study revealed F. nucleatum induction of 14 oncogenic miRNAs, and specifically, 12 miRNAs were linked with CRC.IMPORTANCEOur study investigated oral commensal Fusobacterium nucleatum, a critical bacterium associated with gum disease, adverse pregnancy outcomes, and enriched several tumors, including colorectal cancer (CRC). Recently, microRNAs have emerged as critical players in the interactions between host and microbes, and many host functions have been reported to be regulated by miRNAs during infection. F. nucleatum oral infection in mice induced gum disease, disseminated to the heart, lungs, and several miRNAs. Elevated miR-361 expression was linked to multiple cancers. In addition, miR-126-5p expression has been reported as a potential biomarker in patients with periodontitis and coronary artery disease, indicating F. nucleatum's virulence potential. The 13 miRNAs induced by F. nucleatum are linked to 13 multiple malignancies, including CRC. These results indicate that F. nucleatum acts as a potent cancer-causing bacterium. This study opens new avenues for exploring F. nucleatum's role in gum disease and its link with cancer.
Keywords: CRC miRNAs; F. nucleatum; machine learning models; miRNAs; periodontal disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Altered microRNA expression correlates with reduced TLR2/4-dependent periodontal inflammation and bone resorption induced by polymicrobial infection.Microbiol Spectr. 2025 Aug 14:e0016025. doi: 10.1128/spectrum.00160-25. Online ahead of print. Microbiol Spectr. 2025. PMID: 40810551
-
Altered microRNA Expression Correlates with Reduced TLR2/4-Dependent Periodontal Inflammation and Bone Resorption Induced by Polymicrobial Infection.bioRxiv [Preprint]. 2025 Jan 11:2025.01.10.632435. doi: 10.1101/2025.01.10.632435. bioRxiv. 2025. Update in: Microbiol Spectr. 2025 Aug 14:e0016025. doi: 10.1128/spectrum.00160-25. PMID: 39829929 Free PMC article. Updated. Preprint.
-
Fusobacterium's link to colorectal neoplasia sequenced: A systematic review and future insights.World J Gastroenterol. 2017 Dec 28;23(48):8626-8650. doi: 10.3748/wjg.v23.i48.8626. World J Gastroenterol. 2017. PMID: 29358871 Free PMC article.
-
The interactions of Fusobacterium nucleatum and Porphyromonas gingivalis with microRNAs: contributions to oral squamous cell carcinoma.Mol Biol Rep. 2025 Aug 12;52(1):821. doi: 10.1007/s11033-025-10926-0. Mol Biol Rep. 2025. PMID: 40794318
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Krieger M, AbdelRahman YM, Choi D, Palmer EA, Yoo A, McGuire S, Kreth J, Merritt J. 2024. Stratification of Fusobacterium nucleatum by local health status in the oral cavity defines its subspecies disease association. Cell Host Microbe 32:479–488. doi: 10.1016/j.chom.2024.02.010 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

