Regulatory networks of FUR and NtcA are intertwined by transcriptional regulators, two-component systems, serine/threonine kinases, and sigma factors in Anabaena sp. PCC 7120
- PMID: 40558093
- PMCID: PMC12282138
- DOI: 10.1128/msystems.00373-25
Regulatory networks of FUR and NtcA are intertwined by transcriptional regulators, two-component systems, serine/threonine kinases, and sigma factors in Anabaena sp. PCC 7120
Abstract
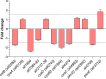
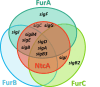
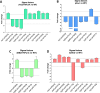
FUR proteins in Anabaena sp. PCC 7120 (FurA/Fur, FurB/Zur, and FurC/PerR) are a family of transcriptional regulators involved in the control of highly important metabolic processes such as the maintenance of metal homeostasis, the regulation of oxidative stress response, and the adaptation to nitrogen starvation. Previous RNAseq analyses of FUR misregulation strains revealed a broad panel of genes directly modulated by these transcriptional regulators. However, the expression of several regulatory proteins was also altered, indicating that FUR proteins could extend their influence by exerting a second level of regulation through some members of their regulons. In this work, by combining differential gene expression data and electrophoretic mobility shift assays (EMSAs), we sought to identify novel direct targets of FUR proteins with regulatory functions, namely, transcriptional regulators, two-component systems, sigma factors, and other proteins with regulatory functions such as serine/threonine kinases. This allowed us to build a network composed of these regulatory proteins that are directly modulated by FUR proteins. In addition, taking into account the role of FUR proteins in the regulation of nitrogen metabolism, the overlap between FUR and NtcA regulatory networks was studied, revealing that an important part of the FUR network is coregulated by NtcA. These results unveil a complex network in Anabaena in which regulatory proteins hierarchically below FUR or NtcA proteins could be controlling the expression of several genes, connecting the integration of stress signaling performed by FUR and NtcA to a wide set of cyanobacterial transcriptional responses.IMPORTANCEFUR proteins in Anabaena sp. PCC 7120 are a family of global transcriptional regulators that control several cellular processes ranging from metal homeostasis to nitrogen metabolism. Apart from directly regulating their target genes, the differential expression of several regulatory genes in RNAseq analyses of FUR misregulation strains suggests that these transcriptional regulators could also control the expression of many targets indirectly. Here, we report that FUR proteins from Anabaena sp. PCC 7120 directly modulate the expression of transcriptional regulators, two-component systems, sigma factors, and serine/threonine kinases, revealing that these regulators indirectly modulate a wide number of genes and cellular processes through this regulatory network. Besides, it was found that an important part of this network is co-regulated by NtcA, connecting the integration of FUR and NtcA stress signals and suggesting that the FUR regulatory network could be involved in the adaptive responses to nitrogen deficiency.
Keywords: Anabaena sp. PCC 7120; FUR (ferric uptake regulator); NtcA; regulatory networks; serine/threonine kinase; sigma factor; transcriptional regulator; two-component system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
