Three-Dimensional Modeling of Camelus dromedarius T Cell Receptor Gamma (TRG)_Delta (TRD)/CD1D Complex Reveals Different Binding Interactions Depending on the TRD CDR3 Length
- PMID: 40558100
- PMCID: PMC12189835
- DOI: 10.3390/antib14020046
Three-Dimensional Modeling of Camelus dromedarius T Cell Receptor Gamma (TRG)_Delta (TRD)/CD1D Complex Reveals Different Binding Interactions Depending on the TRD CDR3 Length
Abstract
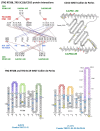
Background: In the adaptive immune response of the dromedary (Camelus dromedarius, Camdro), the T cell receptor (TR) repertoire of the gamma-delta (γδ) T cells is unusually diversified both by somatic hypermutation in rearranged TR gamma (TRG) and delta (TRD) genes and by the diversity in sequence and length of the third complementarity-determining region (CDR3) of the TRD chain.
Methods: The purpose was to investigate, in the absence of 3D structures, the role of Camdro γδ T cells, focusing on the binding interactions at the interface between the V-gamma and V-delta domains, and in complex with the CD1D, a major histocompatibily class I (MH1)-like glycoprotein presenting lipid antigen in association with B2M. A combination of hypermutated TRG dromedary cDNA clones was paired with TRD clones bearing very long, long, or short CDR3s, all isolated from the spleen of a single animal.
Results: The 3D models of the Camdro TRG_TRD/CD1D_B2M complexes were inferred using the Homo sapiens 3D structure and the ImMunoGeneTics (IMGT) numbering for V, C, and G domains, and investigated for binding interactions at the interface of the paired V-gamma_V-delta and at the interface with CD1D. Our results suggest that transcripts with long CDR3s may derive from a population of CD1D-restricted γδ T cells. Both the CD1D G-alpha1-like and G-alpha-2 like domain helices were contacted by both the V-gamma and V-delta CDR-IMGT loops.
Conclusions: Our findings further emphasize the similarity between the γδ T cells population we analyzed in Camelus dromedarius and the CD1D-restricted γδ NKT cells in Homo sapiens.
Keywords: 3D modeling; Camelus dromedarius; IMGT unique numbering; T cell receptor; TRG and TRD loci; multiple sequence alignments (MSA); protein–protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

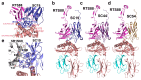
Similar articles
-
3D structures inferred from cDNA clones identify the CD1D-Restricted γδ T cell receptor in dromedaries.Front Immunol. 2022 Aug 9;13:928860. doi: 10.3389/fimmu.2022.928860. eCollection 2022. Front Immunol. 2022. PMID: 36016959 Free PMC article.
-
Characteristics of the somatic hypermutation in the Camelus dromedarius T cell receptor gamma (TRG) and delta (TRD) variable domains.Dev Comp Immunol. 2014 Oct;46(2):300-13. doi: 10.1016/j.dci.2014.05.001. Epub 2014 May 13. Dev Comp Immunol. 2014. PMID: 24836674
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Psychological therapies for treatment-resistant depression in adults.Cochrane Database Syst Rev. 2018 May 14;5(5):CD010558. doi: 10.1002/14651858.CD010558.pub2. Cochrane Database Syst Rev. 2018. PMID: 29761488 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
References
-
- Lefranc M.-P., Lefranc G. The Immunoglobulin Factsbook. Academic Press; Cambridge, MA, USA: 2001.
-
- Lefranc M.-P., Lefranc G. The T Cell Receptor FactsBook. Academic Press; London, UK: 2001.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

