Harnessing Natural Product Compounds to Target Dormancy Survival Regulator (DosR) in Latent Tuberculosis Infection (LTBI): An In Silico Strategy Against Dormancy
- PMID: 40558118
- PMCID: PMC12190169
- DOI: 10.3390/arm93030019
Harnessing Natural Product Compounds to Target Dormancy Survival Regulator (DosR) in Latent Tuberculosis Infection (LTBI): An In Silico Strategy Against Dormancy
Abstract
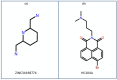
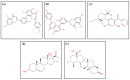
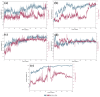
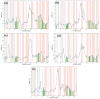
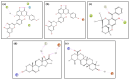
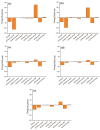
Dormancy occurs when Mycobacterium tuberculosis (Mtb) enters a non-replicating and metabolically inactive state in response to hostile environment. During this state, it is highly resistant to conventional antibiotics, which increase the urgency to develop new potential drugs against dormant bacilli. In view of this, the dormancy survival regulator (DosR) protein is thought to be an essential component that plays a key role in bacterial adaptation to dormancy during hypoxic conditions. Herein, the NP-lib database containing natural product compounds was screened virtually against the binding site of the DosR protein using the MTiopen screen web server. A series of computational analyses were performed, including redocking, intermolecular interaction analysis, and MDS, followed by binding free energy analysis. Through screening, 1000 natural product compounds were obtained with docking energy ranging from -8.5 to -4.1 kcal/mol. The top four lead compounds were then selected for further investigation. On comparative analysis of intermolecular interaction, dynamics simulation and MM/GBSA calculation revealed that M3 docked with the DosR protein (docking score = -8.1 kcal/mol, RMSD = ~7 Å and ΔG Bind = -53.51 kcal/mol) exhibited stronger stability than reference compound Ursolic acid (docking score = -6.2 kcal/mol, RMSD = ~13.5 Å and ΔG Bind = -44.51 kcal/mol). Hence, M3 is recommended for further validation through in vitro and in vivo studies against latent tuberculosis infection.
Keywords: DosR; dormancy; molecular dynamics simulation; structure-based virtual screening; tuberculosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Exploring Type II Diabetes Inhibitors from Genus Daphne Plant-species: An Integrated Computational Study.Comb Chem High Throughput Screen. 2025;28(8):1413-1442. doi: 10.2174/0113862073262227231005074024. Comb Chem High Throughput Screen. 2025. PMID: 38584562
-
Suppressing lipid biosynthesis in Mycobacterium tuberculosis through polyketide synthase 13 thioesterase inhibition: Insights from computational analysis.J Infect Public Health. 2025 Sep;18(9):102835. doi: 10.1016/j.jiph.2025.102835. Epub 2025 May 26. J Infect Public Health. 2025. PMID: 40460691
-
Computational Investigation of Phytochemicals Targeting Isocitrate Lyase to Inhibit Mycobacterium tuberculosis.Curr Drug Discov Technol. 2025 Jun 16. doi: 10.2174/0115701638364461250603050239. Online ahead of print. Curr Drug Discov Technol. 2025. PMID: 40530732
-
Accurate diagnosis of latent tuberculosis in children, people who are immunocompromised or at risk from immunosuppression and recent arrivals from countries with a high incidence of tuberculosis: systematic review and economic evaluation.Health Technol Assess. 2016 May;20(38):1-678. doi: 10.3310/hta20380. Health Technol Assess. 2016. PMID: 27220068 Free PMC article.
-
Xpert® MTB/RIF assay for extrapulmonary tuberculosis and rifampicin resistance.Cochrane Database Syst Rev. 2018 Aug 27;8(8):CD012768. doi: 10.1002/14651858.CD012768.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2021 Jan 15;1:CD012768. doi: 10.1002/14651858.CD012768.pub3. PMID: 30148542 Free PMC article. Updated.
References
-
- Wu Y., Xiong Y., Zhong Y., Liao J., Wang J. Role of Dormancy Survival Regulator and Resuscitation-Promoting Factors Antigens in Differentiating between Active and Latent Tuberculosis: A Systematic Review and Meta-Analysis. BMC Pulm. Med. 2024;24:541. doi: 10.1186/s12890-024-03348-4. - DOI - PMC - PubMed
-
- Global Tuberculosis Report. 2024. [(accessed on 29 May 2025)]. Available online: https://www.who.int/teams/global-programme-on-tuberculosis-and-lung-heal....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

