The Genetic Background and Culture Medium Only Marginally Affect the In Vitro Evolution of Pseudomonas aeruginosa Toward Colistin Resistance
- PMID: 40558191
- PMCID: PMC12189927
- DOI: 10.3390/antibiotics14060601
The Genetic Background and Culture Medium Only Marginally Affect the In Vitro Evolution of Pseudomonas aeruginosa Toward Colistin Resistance
Abstract
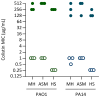
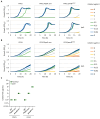
Background/Objectives: Colistin is a last-resort treatment for Pseudomonas aeruginosa multidrug-resistant infections, but resistance to it is emerging. While colistin resistance in P. aeruginosa is typically associated with chromosomal mutations inducing lipopolysaccharide (LPS) aminoarabinosylation, other mutations unrelated to LPS modifications have been proposed to influence the extent of colistin resistance. Here, we examined whether the genetic background and culture conditions affect the evolution of high-level colistin resistance in this bacterium. Methods: We performed in vitro evolution experiments in the presence or absence of increasing colistin concentrations with two phylogenetically distant reference strains in a standard laboratory medium and in two media mimicking P. aeruginosa growth during lung or systemic infections. Resistance-associated mutations were identified by comparative genomics, and the role of selected mutated genes was validated by allele replacement, deletion, or conditional mutagenesis. Results: Most colistin-resistant mutants carried mutations in genes belonging to four functional groups: two-component systems controlling LPS aminoarabinosylation (PmrAB, PhoPQ), LPS biosynthesis, the production of the polyamine norspermidine, and fatty acid metabolism. No mutation was exclusively and invariably associated with a specific strain or medium. We demonstrated that norspermidine is detrimental to the acquisition of colistin resistance upon PmrAB activation and that impaired fatty acid biosynthesis can promote colistin resistance, even if it increases susceptibility to other antibiotics. Conclusions: The evolution of colistin resistance in P. aeruginosa appeared to be only marginally affected by the genetic background and culture conditions. Notably, mutations in fatty acid biosynthetic genes represent a newly identified genetic determinant of P. aeruginosa colistin resistance, warranting further investigation in clinical isolates.
Keywords: PA14; PAO1; artificial sputum medium; colistin; fatty acids; human serum; in vitro evolution; lipid A; norspermidine; polymyxins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2017 Apr 25;4(4):CD004197. doi: 10.1002/14651858.CD004197.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Jun 2;6:CD004197. doi: 10.1002/14651858.CD004197.pub6. PMID: 28440853 Free PMC article. Updated.
-
Comparative evaluation of colistin broth disc elution (CBDE) and broth microdilution (BMD) in clinical isolates of Pseudomonas aeruginosa with special reference to heteroresistance.Indian J Med Microbiol. 2024 Jan-Feb;47:100494. doi: 10.1016/j.ijmmb.2023.100494. Epub 2023 Oct 25. Indian J Med Microbiol. 2024. PMID: 37890411
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
The fatty acid synthesis gene fabY affects lipopolysaccharide synthesis and colistin susceptibility in Pseudomonas aeruginosa.Microbiol Spectr. 2025 Aug 5;13(8):e0133925. doi: 10.1128/spectrum.01339-25. Epub 2025 Jul 15. Microbiol Spectr. 2025. PMID: 40662729 Free PMC article.
-
Xpert® MTB/RIF assay for extrapulmonary tuberculosis and rifampicin resistance.Cochrane Database Syst Rev. 2018 Aug 27;8(8):CD012768. doi: 10.1002/14651858.CD012768.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2021 Jan 15;1:CD012768. doi: 10.1002/14651858.CD012768.pub3. PMID: 30148542 Free PMC article. Updated.
References
-
- Narimisa N., Keshtkar A., Dadgar-Zankbar L., Bostanghadiri N., Far Y.R., Shahroodian S., Zahedi Bialvaei A., Razavi S. Prevalence of colistin resistance in clinical isolates of Pseudomonas. aeruginosa: A systematic review and meta-analysis. Front. Microbiol. 2024;15:1477836. doi: 10.3389/fmicb.2024.1477836. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

