Spatiotemporal Adaptations-Driven Dynamic Thra Activation Simulates a Skin Wound Healing Response
- PMID: 40558382
- PMCID: PMC12442676
- DOI: 10.1002/advs.202506651
Spatiotemporal Adaptations-Driven Dynamic Thra Activation Simulates a Skin Wound Healing Response
Abstract
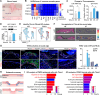
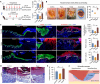
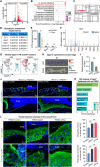
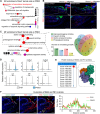
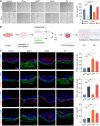
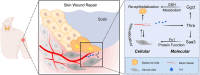
The evolutionary adaptation of skin repair drives sequential regenerative phases: epidermal proliferation rapidly restores barrier function, followed by dermal reconstruction through extracellular matrix remodeling to establish structural support, yet the molecular coordination of this spatiotemporal program remains unclear. While the endocrine system is crucial in modulating wound repair, the critical hormone receptors orchestrating tissue-layer-specific responses are unidentified. Here, bulk and single-cell RNA sequencing, spatial transcriptomics, and in vivo/in vitro analyses in mouse models of hyperthyroidism and hypothyroidism, as well as wound and skin organoid models, are employed to identify the thyroid hormone receptor Thra as a key regulator of phase-coupled regeneration through two distinct yet coordinated mechanisms. In the initial phase, epidermal Thra activates glutathione metabolism via Gamma-Glutamylcyclotransferase (GGCT), driving keratin filament assembly to accelerate reepithelialization. In the subsequent phase, dermal Thra mediates the Serum Amyloid A3 (SAA3)-Fibronectin 1 (FN1) interaction, establishing angiogenic niches essential for matrix maturation. Using the self-assembled epidermis-dermis dynamic skin organoid model, Thra's role in simulating the wound healing process is further confirmed. This study highlights the essential role of spatiotemporal adaptability in wound repair using Thra as a paradigm and provides insights for developing clinical strategies to enhance skin wound healing.
Keywords: SAA3; glutathione metabolism; skin organoids; thyroid hormone; wound healing.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ben‐Moshe S., Veg T., Manco R., Dan S., Papinutti D., Lifshitz A., Kolodziejczyk A. A., Bahar Halpern K., Elinav E., Itzkovitz S., Cell Stem Cell 2022, 29, 973. - PubMed
-
- Chen F., Tchorz J. S., Cell Stem Cell 2022, 29, 871. - PubMed
-
- Peña O. A., Martin P., Nat. Rev. Mol. Cell Biol. 2024, 25, 599. - PubMed
MeSH terms
Grants and funding
- 82373509/National Natural Science Foundation of China
- 2023YFC2508200/National Key Research and Development Program of China
- 2023CDJKYJH066/Fundamental Research Funds for the Central Universities
- 2022CDJYGRH-003/Fundamental Research Funds for the Central Universities
- 2023CDJXY-050/Fundamental Research Funds for the Central Universities
- 2024CDJXY017/Fundamental Research Funds for the Central Universities
- cstc2021ycjh-bgzxm0197/Chongqing Talents: Exceptional Young Talents Project
- 02210011044110/Scientific Research Foundation from Chongqing University
- H20231432/Research Horizontal Projects of Chongqing University
- H20230936/Research Horizontal Projects of Chongqing University
- 005884-00001/CMU-USC
LinkOut - more resources
Full Text Sources
Miscellaneous
