Genomic Characterization of Linezolid-Resistant Clostridioides difficile Harboring cfr Variants
- PMID: 40558391
- PMCID: PMC12190932
- DOI: 10.3390/biotech14020042
Genomic Characterization of Linezolid-Resistant Clostridioides difficile Harboring cfr Variants
Abstract
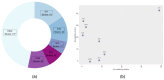
The emergence of antimicrobial resistance (AMR) in Clostridium difficile (C. difficile), particularly to last-line antibiotics such as linezolid, represents a critical challenge in clinical settings. This study investigates the genomic epidemiology of linezolid-resistant C. difficile, focusing on the distribution and mutational patterns of the chloramphenicol-florfenicol resistance (cfr) gene and its association with multidrug resistance. We analyzed 514 clinical isolates (354 from NCBI Pathogen Detection, 160 from EnteroBase), revealing distinct prevalence patterns among cfr subtypes: cfr(C) was dominant (156/354 NCBI strains; 101/160 EnteroBase strains), whereas cfr(B) frequently harbored missense mutations (p.R247K, p.V294I, and less commonly p.A334T). The cfr(E) subtype was exclusively identified in ribotype 027 (RT027) strains. Notably, cfr(C) exhibited a strong association with RT017, correlating with a conserved 99 bp genomic deletion. Phylogenetic analysis linked cfr-carriage to predominant sequence types (ST1 in NCBI strains, ST37 in EnteroBase isolates). Furthermore, the co-occurrence of cfr with additional AMR genes conferred resistance to macrolides (erythromycin, azithromycin) and tetracyclines, indicating a convergent evolution toward multidrug resistance. These findings underscore the interplay between cfr mutations, hypervirulent ribotypes, and AMR dissemination, necessitating enhanced surveillance to mitigate the spread of resistant C. difficile lineages.
Keywords: C. difficile; Cfr subtypes; ST; antimicrobial resistance genes; linezolid.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

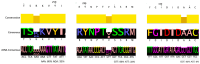

References
LinkOut - more resources
Full Text Sources
