The Use of CRISPR-Cas Systems for Viral Detection: A Bibliometric Analysis and Systematic Review
- PMID: 40558461
- PMCID: PMC12190998
- DOI: 10.3390/bios15060379
The Use of CRISPR-Cas Systems for Viral Detection: A Bibliometric Analysis and Systematic Review
Abstract
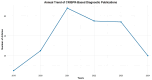
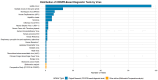
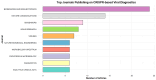
Viral infections impose a significant burden on global public health and the economy. This study examines the current state of CRISPR-Cas system research, focusing on their applications in viral detection and their evolution over recent years. A bibliometric analysis and systematic review were conducted using articles published between 2019 and 2024, retrieved from Web of Science, Scopus, and PubMed databases. Out of 2713 identified articles, 194 were included in the analysis. The findings reveal substantial growth in scientific output related to CRISPR-Cas systems, with the United States leading in research and development in this field. The rapid increase in CRISPR-Cas research during this period underscores its immense potential to transform viral diagnostics. With advantages such as speed, precision, and suitability for deployment in resource-limited settings, CRISPR-Cas systems outperform many traditional diagnostic methods. The concerted efforts of scientists worldwide further highlight the promising future of this technology. CRISPR-Cas systems are emerging as a powerful alternative, offering the possibility of expedited and accessible point-of-care testing and paving the way for more equitable and effective diagnostics on a global scale.
Keywords: CRISPR-Cas systems; bibliometric analysis; isothermal amplification; viral diagnostic assay.
Conflict of interest statement
The authors declare no competing interests.
Figures
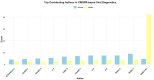
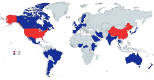
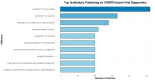
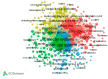
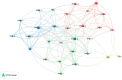
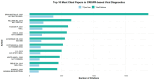
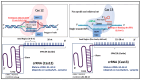

References
-
- FDA Coronavirus (COVID-19) Update: FDA Alerts Consumers About Unauthorized Fraudulent COVID-19 Test Kits. [(accessed on 24 September 2024)]; Available online: https://fdareporter.com/stories/fda-coronavirus-covid-19-update-fda-aler...
-
- Radio-Canada En C.-B. 30 % des Tests de Dépistage Donnent un Faux Résultat Négatif à la COVID-19|COVID-19: Tout Sur la Pandémie. [(accessed on 24 September 2024)]. Available online: https://ici.radio-canada.ca/nouvelle/1693947/covid-19-faux-tests-resulta....
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

