Regulation of Myogenesis by MechanomiR-200c/FoxO3 Axis
- PMID: 40558495
- PMCID: PMC12191418
- DOI: 10.3390/cells14120868
Regulation of Myogenesis by MechanomiR-200c/FoxO3 Axis
Abstract
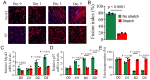
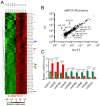
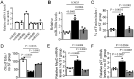
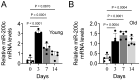
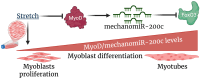
Cyclic mechanical stretch has been shown to inhibit myoblast differentiation while promoting proliferation. However, the underlying molecular mechanisms are not well understood. Here, we report that mechanical stretch inhibits the differentiation of mouse primary myoblasts by promoting the cell cycle program and by inhibiting the expression of the myogenic regulator MyoD. Stretch alters the miRNA expression profile as evidenced by miRNA microarray analysis. We identified miR-200c as one of the highly downregulated mechanosensitive miRNAs (mechanomiRs) whose expression level was increased during differentiation. This suggests that mechanomiRs-200c is a myogenic miRNA. Overexpression of mechanomiR-200c revoked the effect of stretch on myoblast differentiation, and the introduction of the mechanomiR-200c antagomir restored the stretch effect. This suggests that stretch blocks differentiation, in part, through mechanomiR-200c. The gene encoding the transcription factor FoxO3 is a known direct target of mechanomiR-200c. Interestingly, MyoD binds to the mechanomiR-200c promoter in differentiating myoblasts, whereas stretch appears to reverse such binding. Our data further demonstrate that the levels of mechanomiR-200c are robustly elevated during the early stage of the muscle repair process in young mice, but not in the injured muscle of aged mice. Overall, we identified a novel pathway, MyoD/mechanomiR-200c/FoxO3a, and the potential mechanism by which stretch inhibits myoblast differentiation.
Keywords: MyoD; differentiation; mechanical stretch; microRNAs; myoblasts.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
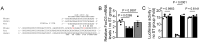


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

