Epigenetic Reprogramming of Cell Identity in the Rat Primary Neuron-Glia Cultures Involves Histone Serotonylation
- PMID: 40558532
- PMCID: PMC12191383
- DOI: 10.3390/cells14120905
Epigenetic Reprogramming of Cell Identity in the Rat Primary Neuron-Glia Cultures Involves Histone Serotonylation
Abstract
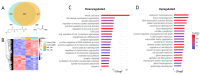
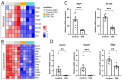
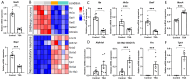
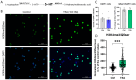
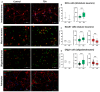
Epigenetic rearrangements can create a favorable environment for the intrinsic plasticity of brain cells, leading to cellular reprogramming into virtually any cell type through the induction of cell-specific transcriptional programs. In this study, we assessed how chromatin remodeling induced by broad-spectrum HDAC inhibitors affects cellular differentiation trajectories in rat primary neuron-glia cultures using a combination of transcriptomics, qPCR, and cytochemistry. We described the epigenetic regulation of transcriptional programs controlled by master transcription factors and neurotrophins in the context of neuronal and glial differentiation and evaluated the expression of representative cell-specific markers. The results obtained suggest that HDAC inhibitors reduce the proliferative potential of cultured cells and induce transcriptomic changes associated with cell differentiation and specialization. Particularly, we revealed a significant upregulation of genes typically expressed in neuromodulatory neurons and the downregulation of genes expressed in glia and inhibitory neurons. Transcriptional changes were accompanied by continuous elevation of histone serotonylation levels in both neurons and glia. Emerging shortly after HDAC inhibition, a complex chromatin remodeling, which includes histone serotonylation, persists over many hours in distinct brain cells. We assume that this sustained epigenetic mechanism likely helps to maintain transcriptional changes associated with cell fate commitment, possibly priming cells for long-term fate conversion.
Keywords: HDAC; cell identity; epigenetics; glia; histone serotonylation; neuron; reprogramming; transcriptional program.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Humphrey G.W., Wang Y.H., Hirai T., Padmanabhan R., Panchision D.M., Newell L.F., McKay R.D., Howard B.H. Complementary Roles for Histone Deacetylases, 1, 2, and 3 in Differentiation of Pluripotent Stem Cells. Differentiation. 2008;76:348–356. doi: 10.1111/j.1432-0436.2007.00232.x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

