Transparent 3-Layered Bacterial Nanocellulose as a Multicompartment and Biomimetic Scaffold for Co-Culturing Cells
- PMID: 40558895
- PMCID: PMC12194138
- DOI: 10.3390/jfb16060208
Transparent 3-Layered Bacterial Nanocellulose as a Multicompartment and Biomimetic Scaffold for Co-Culturing Cells
Abstract
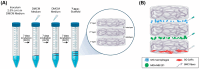
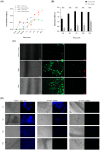
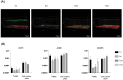
Three-dimensional (3D) cell culture models are widely used to provide a more physiologically relevant microenvironment in which to host and study desired cell types. These models vary in complexity and cost, ranging from simple and inexpensive to highly sophisticated and costly systems. In this study, we introduce a novel translucent multi-compartmentalized stacked multilayered nanocellulose scaffold and describe its fabrication, characterization, and potential application for co-culturing multiple cell types. The scaffold consists of bacterial nanocellulose (BNC) layers separated by interlayers of a lower density of nanocellulose fibers. Using this system, we co-cultured the MDA-MB-231 cell line with two tumor-associated cell types, namely BC-CAFs and M2 macrophages, to simulate the tumor microenvironment (TME). Cells remained viable and metabolically active for up to 15 days. Confocal microscopy showed no signs of cell invasion. However, BC-CAFs and MDA-MB-231 cells were frequently observed within the same layer. The expression of breast cancer-related genes was analyzed to assess the downstream functionality of the cells. We found that the E-cadherin expression was 20% lower in cancer cells co-cultured in the multi-compartmentalized scaffold than in those cultured in 2D plates. Since E-cadherin plays a critical role in preventing the initial dissociation of epithelial cells from the primary tumor mass and is often downregulated in the tumor microenvironment in vivo, this finding suggests that our scaffold more effectively recapitulates the complexity of a tumor microenvironment.
Keywords: 3D cell culture model; bacterial nanocellulose; triple-cell co-culture.
Conflict of interest statement
All authors declare that they have no conflicts of interest. EA.hy929 experiments were performed at EMBRAPA, under approval of A.P.A.B. The company has no conflict of interest on this work.
Figures

Similar articles
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

