Integrated Transcriptomics and Metabolomics Provide Insight into Degeneration-Related Molecular Mechanisms of Morchella importuna During Repeated Subculturing
- PMID: 40558932
- PMCID: PMC12194730
- DOI: 10.3390/jof11060420
Integrated Transcriptomics and Metabolomics Provide Insight into Degeneration-Related Molecular Mechanisms of Morchella importuna During Repeated Subculturing
Abstract
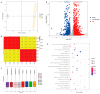
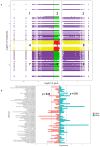
This study investigated Morchella importuna strain degeneration during repeated subculturing and employed metabolomics, transcriptomics, and other techniques to explore its molecular mechanisms. Significant metabolic and transcriptional differences were observed between normal mycelia (NM) and degenerated mycelia (DG). Metabolomic analysis revealed 699 differentially expressed metabolites (DEMs) that were predominantly enriched in secondary metabolite biosynthesis pathways, particularly flavonoids and indole alkaloids. Total flavonoid content was markedly higher in NM than in DG, with most flavonoid compounds showing reduced levels in degenerated strains. Transcriptomic profiling revealed 2691 differentially expressed genes (DEGs), primarily associated with metabolic pathways and genetic information processing. Integrated analysis showed that metabolic dynamics were regulated by DEGs, with pyruvate metabolism being significantly enriched. The FunBGCeX tool identified biosynthetic gene clusters (BGCs) in the M. importuna genome, highlighting the critical role of the non-reducing polyketide synthase (NR-PKS) gene in flavonoid biosynthesis. This gene exhibited significantly downregulated expression in DG strains. These findings indicate that M. importuna degeneration resulted from systemic dysregulation of gene expression networks and metabolic pathway reorganization. The results presented herein also provide theoretical insights into degeneration mechanisms and potential prevention strategies for this edible fungus.
Keywords: Morchella importuna; mechanisms; non-reducing polyketide synthase; strain degeneration.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

