Telomere-to-Telomere Assembly of the Cordyceps militaris CH1 Genome and Integrated Transcriptomic and Metabolomic Analyses Provide New Insights into Cordycepin Biosynthesis Under Light Stress
- PMID: 40558975
- PMCID: PMC12194794
- DOI: 10.3390/jof11060461
Telomere-to-Telomere Assembly of the Cordyceps militaris CH1 Genome and Integrated Transcriptomic and Metabolomic Analyses Provide New Insights into Cordycepin Biosynthesis Under Light Stress
Abstract
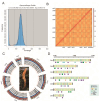
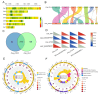
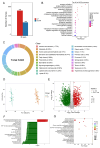
Cordyceps militaris, a model species in the genus Cordyceps, is widely distributed globally and is known for its significant medicinal value. It has been traditionally used in Chinese medicine to enhance immunity, alleviate fatigue, and treat tumors, among other therapeutic purposes. Here, we successfully assembled a telomere-to-telomere (T2T) level genome of C. militaris CH1 using PacBio HiFi and Hi-C technologies. The assembled genome is 32.67 Mb in size, with an N50 of 4.70 Mb. Gene prediction revealed a total of 10,749 predicted genes in the C. militaris CH1 genome, with a gene completeness of 99.20%. Phylogenetic analysis showed the evolutionary relationship between C. militaris CH1 and other Cordyceps species, suggesting that the divergence between this strain and C. militaris ATCC 34164 occurred approximately 1.36 Mya. Combined transcriptomic and metabolomic analyses identified 842 differentially expressed genes and 2052 metabolites that were significantly altered under light stress, primarily involving key pathways related to amino acid metabolism, purine metabolism, and secondary metabolite biosynthesis. Joint analysis of genes and metabolites revealed 79 genes coding for enzymes associated with the synthesis of adenine and adenosine, with the expression of 52 genes being upregulated, consistent with the accumulation trends of adenine and adenosine. Four gene clusters related to the synthesis of cordycepin were identified, with a significant upregulation of cns3 (FUN_003263), suggesting that light stress may promote cordycepin biosynthesis. This comprehensive analysis not only provides new insights into the genomics, metabolomics, and functional gene research of C. militaris CH1 but also offers a potential biological foundation for understanding the synthesis mechanisms of cordycepin and its efficient production.
Keywords: Cordyceps militaris CH1; comparative genomics; cordycepin biosynthesis; phylogenetic evolution; telomere-to-telomere; transcriptome and metabolome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Integrating Network Pharmacology and Optimization of Preparation Methods to Enhance the Anticancer Effect of Cordyceps militaris on Lung Cancer.Anticancer Res. 2025 Jul;45(7):2963-2984. doi: 10.21873/anticanres.17663. Anticancer Res. 2025. PMID: 40578945
-
The chromosome-level genome of Hemerocallis middendorffii provides new insights into the floral scents and color biosynthesis in Chinese native daylily.BMC Plant Biol. 2025 Jul 4;25(1):874. doi: 10.1186/s12870-025-06863-6. BMC Plant Biol. 2025. PMID: 40615786 Free PMC article.
-
Effects of cottonseed powder and perilla oil on cordycepin production from Cordyceps militaris in liquid culture.Braz J Microbiol. 2025 Sep;56(3):1495-1504. doi: 10.1007/s42770-025-01713-x. Epub 2025 Jun 23. Braz J Microbiol. 2025. PMID: 40549333
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Single-incision sling operations for urinary incontinence in women.Cochrane Database Syst Rev. 2017 Jul 26;7(7):CD008709. doi: 10.1002/14651858.CD008709.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Oct 27;10:CD008709. doi: 10.1002/14651858.CD008709.pub4. PMID: 28746980 Free PMC article. Updated.
References
-
- Shrestha B., Zhang W., Zhang Y., Liu X. The medicinal fungus Cordyceps militaris: Research and development. Mycol. Prog. 2012;11:599–614. doi: 10.1007/s11557-012-0825-y. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

