Comparative IP-MS Reveals HSPA5 and HSPA8 Interacting with Hemagglutinin Protein to Promote the Replication of Influenza A Virus
- PMID: 40559543
- PMCID: PMC12195859
- DOI: 10.3390/pathogens14060535
Comparative IP-MS Reveals HSPA5 and HSPA8 Interacting with Hemagglutinin Protein to Promote the Replication of Influenza A Virus
Abstract
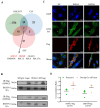
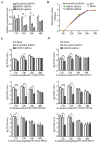
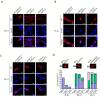
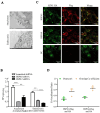
The influenza A viruses (IAV) are the principal pathogens for annual (seasonal) influenza, which cause world-wide outbreaks in poultry and pose a persistent threat to public health. The Hemagglutinin protein (HA) of IAV promotes virus infection by binding the host membrane receptor and mediating virus-host membrane fusion. Immunoprecipitation-mass spectrometry (IP-MS) provides global insights into IAV HA-host protein interactions. However, various experimental conditions might affect the identification of interactions. Here, we performed a serial IP-MS to compare interactors of IAV HA in accidental host human, chicken and reservoir host duck cells. We found that the positive ratio of interactors identified by the IP-MS was improved when the transfected HA plasmid had a similar expression level to HA proteins found in IAV virus infection. Comparing interactors in human, chicken and duck cells, we found that HA-interacting host factors might play a role in the susceptibility of accidental hosts (human and chicken) to IAV infection compared to reservoir hosts (duck). We then focused on the function of two heat shock proteins (HSPA5 and HSPA8), which interacted with IAV HA proteins in all three species (human, chicken and duck). We found that both HSPA5 and HSPA8 promoted the IAV replication by enhancing the viral attachment and internalization. These findings extend our knowledge about the mechanisms of IAV entry to host cells and provide target genes to create chickens resistant to avian influenza.
Keywords: HSPA5 and HSPA8; Hemagglutinin (HA); immunoprecipitation–mass spectrometry (IP-MS); influenza A virus (IAV).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Pleschka S. Overview of influenza viruses. Curr. Top. Microbiol. Immunol. 2013;370:1–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

