Establishment of a Sandwich ELISA for Detection of Pan-Merbecoviruses
- PMID: 40559613
- PMCID: PMC12196402
- DOI: 10.3390/pathogens14060605
Establishment of a Sandwich ELISA for Detection of Pan-Merbecoviruses
Abstract
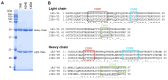
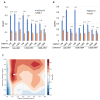
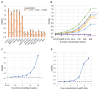
Merbecovirus, a subgenus of Betacoronavirus, includes MERS-CoV and multiple bat-derived viruses with zoonotic potential. Given the unpredictable emergence of these viruses and their genetic diversity, development of broad-spectrum diagnostic tools is expected. In this study, we established a sandwich ELISA targeting the nucleocapsid (N) protein of merbecoviruses. We generated monoclonal antibodies (mAbs) using recombinant N protein of a bat merbecovirus, VsCoV-1, and selected cross-reactive clones for other merbecoviruses. Three mAbs showed strong reactivities with multiple merbecoviruses but not with SARS-CoV-2 or endemic human coronaviruses. Pairwise ELISA screening identified 1A8/10H6 mAbs as the optimal combination for detection of N protein from six merbecoviruses-VsCoV-1, EjCoV-3, MERS-CoV, NeoCoV, HKU4, and HKU5-with limits of detection (LODs) below 7.81 ng/mL, including 1.25 ng/mL for VsCoV-1. Infectious bat merbecovirus EjCoV-3 was detected at 1.3 × 103 PFU/mL. No cross-reactivity was observed with non-merbecoviruses, indicating its high specificity. This sandwich ELISA offers a rapid, reproducible, and cost-effective diagnostic platform with potential for high-throughput screening and automation. Moreover, its design is amenable to adaptation into point-of-care formats such as lateral flow assays, highlighting its value for field-based surveillance and pandemic preparedness.
Keywords: antigen detection; bat merbecoviruses; monoclonal antibody; sandwich ELISA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Designing Sandwich ELISA with Broadly Reactive Anti-Nucleocapsid Monoclonal Antibodies to Detect Bat-Borne Merbecoviruses.Viruses. 2025 Jun 24;17(7):886. doi: 10.3390/v17070886. Viruses. 2025. PMID: 40733504 Free PMC article.
-
In vitro and in vivo characterization of a bat merbecovirus with ACE2- and DPP4-independent cell entry.J Virol. 2025 Jul 22;99(7):e0072725. doi: 10.1128/jvi.00727-25. Epub 2025 Jun 17. J Virol. 2025. PMID: 40525828 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Development of a time-resolved fluorescence-based lateral flow immunoassay for rapid and sensitive diagnosis of Middle East respiratory syndrome.Sci Rep. 2025 Jul 25;15(1):27036. doi: 10.1038/s41598-025-09832-z. Sci Rep. 2025. PMID: 40715253 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
References
-
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.-W., Wong B.H.L., Wong S.S.Y., Leung S.-Y., Chan K.-H., Yuen K.-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

