The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis
- PMID: 40559622
- PMCID: PMC12195753
- DOI: 10.3390/ncrna11030044
The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis
Abstract
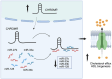
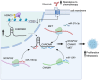
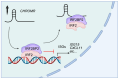
CHROMR is a primate-specific long noncoding RNA with emerging roles in homeostasis and pathophysiology. Elevated blood levels of CHROMR have been observed in patients with cardiovascular disease and several cancers, where it is correlated with poor clinical outcomes. Like many lncRNAs, CHROMR accumulates in both the nucleus and the cytoplasm, and it assumes distinct functions in each of these cellular compartments. In the nucleus, CHROMR sequesters a transcriptional repressor complex to activate interferon-stimulated gene expression and antiviral immunity. In the cytoplasm, CHROMR competitively inhibits microRNAs involved in cholesterol efflux and cell cycle regulation, thereby impacting gene pathways involved in reverse cholesterol transport, HDL biogenesis, and tumor growth. In this review, we detail the multifaceted functions of CHROMR in cholesterol metabolism, innate immunity, and cancer progression. We also explore the potential molecular mechanisms that govern its expression and dynamic subcellular localization, which may be key to its functional versatility. Advancing our understanding of the regulatory networks and cellular environments that shape CHROMR activity will be critical for assessing its promise as a therapeutic target and diagnostic biomarker.
Keywords: CHROMR; cancer; cholesterol metabolism; innate immunity; long noncoding RNA.
Conflict of interest statement
C.v.S. holds a patent on the use of inhibitors targeting
Figures

Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
-
A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection.J Virol. 2017 Mar 29;91(8):e02388-16. doi: 10.1128/JVI.02388-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28148804 Free PMC article.
-
Adenovirus Modulates Toll-Like Receptor 4 Signaling by Reprogramming ORP1L-VAP Protein Contacts for Cholesterol Transport from Endosomes to the Endoplasmic Reticulum.J Virol. 2017 Feb 28;91(6):e01904-16. doi: 10.1128/JVI.01904-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077646 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

