Companion Animals as Reservoirs of Multidrug Resistance-A Rare Case of an XDR, NDM-1-Producing Pseudomonas aeruginosa Strain of Feline Origin in Greece
- PMID: 40559813
- PMCID: PMC12197500
- DOI: 10.3390/vetsci12060576
Companion Animals as Reservoirs of Multidrug Resistance-A Rare Case of an XDR, NDM-1-Producing Pseudomonas aeruginosa Strain of Feline Origin in Greece
Abstract
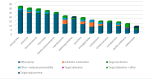
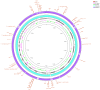
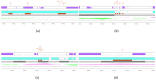
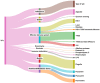
A backyard cat with symptoms of otitis was transferred to a veterinary clinic in Central Greece. A sample was obtained and P. aeruginosa was isolated. The strain exhibited an extensively drug-resistant (XDR) profile, as it was non-susceptible to all tested agents except colistin. DNA extraction and whole-genome sequencing (WGS) were performed using a robotic extractor and Ion Torrent technology, respectively. The genome was assembled and screened for resistance and virulence determinants. The isolate belonged to the high-risk clone ST308 with a total of 67 antibiotic resistance genes (ARGs) and 221 virulence factor-related genes being identified. No plasmids were detected. The metallo-beta-lactamase (MBL) blaNDM-1 gene and 46 efflux pumps were included in the strain's resistome. Both ARGs conferring tolerance to disinfecting agents and biofilm-related genes were identified, associated with the ability of this clone to adapt and persist in healthcare facilities. This case highlights the risk of relevant bacterial clones spreading in the community and even being transmitted to companion animals, causing challenging opportunistic infections to susceptible individuals, while others may become carriers, further spreading the clones to their owners, other animals and the environment.
Keywords: Pseudomonas aeruginosa; ST308; WGS; XDR; blaNDM-1; carbapenemase; cat; virulence factors.
Conflict of interest statement
Authors E.T. and A.T. were employed by Vet Analyseis and author G.A. was employed by CeMIA SA. The companies had no role in the design of the study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The genomic configurations driving antimicrobial resistance and virulence in colistin resistant Pseudomonas aeruginosa from an Egyptian Tertiary Oncology Hospital.PLOS Glob Public Health. 2025 Aug 5;5(8):e0004976. doi: 10.1371/journal.pgph.0004976. eCollection 2025. PLOS Glob Public Health. 2025. PMID: 40763289 Free PMC article.
-
Multidrug-resistant Pseudomonas aeruginosa and its coexistence with β-lactamases at a tertiary care hospital in a low-resource setting: a cross-sectional study with an association of risk factors.Ther Adv Infect Dis. 2025 Jun 18;12:20499361251345920. doi: 10.1177/20499361251345920. eCollection 2025 Jan-Dec. Ther Adv Infect Dis. 2025. PMID: 40538920 Free PMC article.
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2017 Apr 25;4(4):CD004197. doi: 10.1002/14651858.CD004197.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Jun 2;6:CD004197. doi: 10.1002/14651858.CD004197.pub6. PMID: 28440853 Free PMC article. Updated.
-
Variable In Vitro Efficacy of Delafloxacin on Multidrug-Resistant Pseudomonas aeruginosa and the Detection of Delafloxacin Resistance Determinants.Antibiotics (Basel). 2025 May 25;14(6):542. doi: 10.3390/antibiotics14060542. Antibiotics (Basel). 2025. PMID: 40558132 Free PMC article.
-
Delayed antibiotic prescriptions for respiratory infections.Cochrane Database Syst Rev. 2017 Sep 7;9(9):CD004417. doi: 10.1002/14651858.CD004417.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Oct 4;10:CD004417. doi: 10.1002/14651858.CD004417.pub6. PMID: 28881007 Free PMC article. Updated.
References
-
- WHO . WHO Bacterial Priority Pathogens List 2024: Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance. 1st ed. World Health Organization; Geneva, Switzerland: 2024.
LinkOut - more resources
Full Text Sources
Miscellaneous

