Exploring the Role of Bifenthrin in Recurrent Implantation Failure and Pregnancy Loss Through Network Toxicology and Molecular Docking
- PMID: 40559927
- PMCID: PMC12196838
- DOI: 10.3390/toxics13060454
Exploring the Role of Bifenthrin in Recurrent Implantation Failure and Pregnancy Loss Through Network Toxicology and Molecular Docking
Abstract
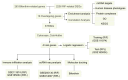
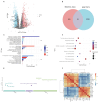
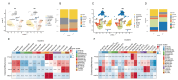
Bifenthrin (BF) is a widely used pyrethroid pesticide recognized as an endocrine-disrupting chemical (EDC). Previous studies have confirmed that chronic exposure to BF is associated with various health risks. However, its potential association with recurrent implantation failure (RIF) and recurrent pregnancy loss (RPL) remains unclear. In this study, the potential targets of BF were identified using several databases, including the Comparative Toxicogenomics Database (CTD), TargetNet, GeneCards, SwissTargetPrediction, and STITCH. Differentially expressed genes (DEGs) associated with RIF were obtained from bulk RNA-seq datasets in the GEO database. Candidate targets were identified by intersecting the predicted BF-related targets with the RIF-associated DEGs, followed by functional enrichment analysis using the DAVID and g:Profiler platforms. Subsequently, hub genes were identified based on the STRING database and Cytoscape. A diagnostic model was then constructed based on these hub genes in the RIF cohort and validated in an independent recurrent pregnancy loss (RPL) cohort. Additionally, we performed single-cell type distribution analysis and immune infiltration profiling based on single-cell RNA-seq and bulk RNA-seq data, respectively. Molecular docking analysis using AutoDock Vina was conducted to evaluate the binding affinity between BF and the four hub proteins, as well as several hormone-related receptors. Functional enrichment results indicated that the candidate genes were mainly involved in apoptotic and oxidative stress-related pathways. Ultimately, four hub genes-BCL2, HMOX1, CYCS, and PTGS2-were identified. The diagnostic model based on these genes exhibited good predictive performance in the RIF cohort and was successfully validated in the RPL cohort. Single-cell transcriptomic analysis revealed a significant increase in the proportion of myeloid cells in RPL patients, while immune infiltration analysis showed a consistent downregulation of M2 macrophages in both RIF and RPL. Moreover, molecular docking analysis revealed that BF exhibited high binding affinity to all four hub proteins and demonstrated strong binding potential with multiple hormone receptors, particularly pregnane X receptor (PXR), estrogen receptor α (ESRα), and thyroid hormone receptors (TR). In conclusion, the association of BF with four hub genes and multiple hormone receptors suggests a potential link to immune and endocrine dysregulation observed in RIF and RPL. However, in vivo and in vitro experimental evidence is currently lacking, and further studies are needed to elucidate the mechanisms by which BF may contribute to RIF and RPL.
Keywords: bifenthrin; endocrine-disrupting chemical; molecular docking; recurrent implantation failure; recurrent pregnancy loss.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

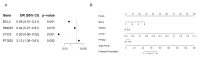

Similar articles
-
Deciphering Shared Gene Signatures and Immune Infiltration Characteristics Between Gestational Diabetes Mellitus and Preeclampsia by Integrated Bioinformatics Analysis and Machine Learning.Reprod Sci. 2025 Jun;32(6):1886-1904. doi: 10.1007/s43032-025-01847-1. Epub 2025 May 15. Reprod Sci. 2025. PMID: 40374866
-
Identification and validation of oxidative stress-related diagnostic markers for recurrent pregnancy loss: insights from machine learning and molecular analysis.Mol Divers. 2025 Aug;29(4):2881-2897. doi: 10.1007/s11030-024-10947-0. Epub 2024 Sep 3. Mol Divers. 2025. PMID: 39225907
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Elucidating the Mechanism of Xiaoqinglong Decoction in Chronic Urticaria Treatment: An Integrated Approach of Network Pharmacology, Bioinformatics Analysis, Molecular Docking, and Molecular Dynamics Simulations.Curr Comput Aided Drug Des. 2025 Jul 16. doi: 10.2174/0115734099391401250701045509. Online ahead of print. Curr Comput Aided Drug Des. 2025. PMID: 40676786
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Ruiz-Alonso M., Blesa D., Diaz-Gimeno P., Gomez E., Fernandez-Sanchez M., Carranza F., Carrera J., Vilella F., Pellicer A., Simon C. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil. Steril. 2013;100:818–824. doi: 10.1016/j.fertnstert.2013.05.004. - DOI - PubMed
-
- Kolanska K., Suner L., Cohen J., Ben Kraiem Y., Placais L., Fain O., Bornes M., Selleret L., Delhommeau F., Feger F., et al. Proportion of Cytotoxic Peripheral Blood Natural Killer Cells and T-Cell Large Granular Lymphocytes in Recurrent Miscarriage and Repeated Implantation Failure: Case-Control Study and Meta-analysis. Arch. Immunol. Ther. Exp. 2019;67:225–236. doi: 10.1007/s00005-019-00546-5. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

