Integrative Spatial Proteomics and Single-Cell RNA Sequencing Unveil Molecular Complexity in Rheumatoid Arthritis for Novel Therapeutic Targeting
- PMID: 40559990
- PMCID: PMC12196869
- DOI: 10.3390/proteomes13020017
Integrative Spatial Proteomics and Single-Cell RNA Sequencing Unveil Molecular Complexity in Rheumatoid Arthritis for Novel Therapeutic Targeting
Abstract
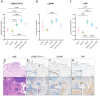
Understanding the heterogeneity of Rheumatoid Arthritis (RA) and identifying therapeutic targets remain challenging using traditional bulk transcriptomics alone, as it lacks the spatial and protein-level resolution needed to fully capture disease and tissue complexities. In this study, we applied Laser Capture Microdissection (LCM) coupled with mass spectrometry-based proteomics to analyze histopathological niches of the RA synovium, enabling the identification of protein expression profiles of the diseased synovial lining and sublining microenvironments compared to their healthy counterparts. In this respect, key pathogenetic RA proteins like membrane proteins (TYROBP, AOC3, SLC16A3, TCIRG1, and NCEH1), and extracellular matrix (ECM) proteins (PLOD2, OGN, and LUM) showed different expression patterns in diseased synovium compartments. To enhance our understanding of cellular dynamics within the dissected regions, we further integrated the proteomic dataset with single-cell RNA sequencing (scRNA-seq), and deduced cell type enrichment, including T cells, fibroblasts, NK cells, myeloid cells, B cells, and synovial endothelial cells. By combining high-resolution spatial proteomics and transcriptomic analyses, we provide novel insights into the molecular mechanisms driving RA, and highlight potential protein targets for therapeutic intervention. This integrative approach offers a more comprehensive view of RA synovial pathology, and mitigates the limitations of traditional bulk transcriptomics in target discovery.
Keywords: extracellular matrix; laser capture microdissection; mass spectrometry; membrane proteins; multi-omics integration; rheumatoid arthritis; scRNA-seq; untargeted proteomics.
Conflict of interest statement
All authors are employed by the AbbVie company. AbbVie funded the study and participated in study design, research, data collection, analysis and interpretation of data, writing, reviewing, and approving the publication. There are no additional conflicts of interest to report.
Figures





References
-
- Nakajima S., Tsuchiya H., Ota M., Ogawa M., Yamada S., Yoshida R., Maeda J., Shirai H., Kasai T., Hirose J., et al. Synovial Tissue Heterogeneity in Japanese Patients with Rheumatoid Arthritis Elucidated Using a Cell-Type Deconvolution Approach. Arthritis Rheumatol. 2023;75:2130–2136. doi: 10.1002/art.42642. - DOI - PubMed
-
- Velickovic M., Fillmore T.L., Attah I.K., Posso C., Pino J.C., Zhao R., Williams S.M., Velickovic D., Jacobs J.M., Burnum-Johnson K.E., et al. Coupling Microdroplet-Based Sample Preparation, Multiplexed Isobaric Labeling, and Nanoflow Peptide Fractionation for Deep Proteome Profiling of the Tissue Microenvironment. Anal. Chem. 2024;96:12973–12982. doi: 10.1021/acs.analchem.4c00523. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

