Unraveling the Central Role of Global Regulator PprI in Deinococcus radiodurans Through Label-Free Quantitative Proteomics
- PMID: 40559992
- PMCID: PMC12197293
- DOI: 10.3390/proteomes13020019
Unraveling the Central Role of Global Regulator PprI in Deinococcus radiodurans Through Label-Free Quantitative Proteomics
Abstract
Background: Deinococcus radiodurans, renowned for its exceptional resistance to radiation, provides a robust model for elucidating cellular stress responses and DNA repair mechanisms. Previous studies have established PprI as a key regulator contributing to radiation resistance through its involvement in DNA damage repair pathways, oxidative stress response, and metabolic regulation.
Methods: Building upon these foundations, our study employs label-free quantitative (LFQ) proteomics coupled with high-resolution mass spectrometry to systematically map pprI deletion protein networks by comparing the global proteomic profiles of pprI knockout and wild-type D. radiodurans strains.
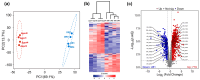
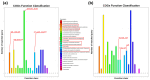
Results: Under stringent screening criteria, we identified 719 significantly higher and 281 significantly lower abundant proteins in the knockout strain compared to wild-type strains. Functional analysis revealed that PprI deficiency disrupts homologous recombination (HR) repair, activates nucleotide excision repair (NER) and base excision repair (BER) as a compensatory mechanism, and impairs Mn/Fe homeostasis and carotenoid biosynthesis, leading to increased oxidative stress. Furthermore, PprI deficiency induces significant metabolic reprogramming, including impaired purine synthesis, compromised cell wall integrity, etc. Conclusions: These proteomic findings delineate the extensive regulatory network influenced by PprI, revealing coordinated perturbations across multiple stress response systems when PprI is absent.
Keywords: DNA damage repair; antioxidant defense; label-free quantitative proteomics; metabolic regulation; radiation response metalloprotease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Makarova K.S., Aravind L., Wolf Y.I., Tatusov R.L., Minton K.W., Koonin E.V., Daly M.J. Genome of the Extremely Radiation-Resistant Bacterium Deinococcus radiodurans Viewed from the Perspective of Comparative Genomics. Microbiol. Mol. Biol. Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. - DOI - PMC - PubMed
-
- White O., Eisen J.A., Heidelberg J.F., Hickey E.K., Peterson J.D., Dodson R.J., Haft D.H., Gwinn M.L., Nelson W.C., Richardson D.L., et al. Genome Sequence of the Radioresistant Bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. - DOI - PMC - PubMed
-
- Yan S., Gao S.S., Zhou P.K. Multi-Functions of Exonuclease 1 in DNA Damage Response and Cancer Susceptibility. Radiat. Med. Prot. 2021;2:146–154. doi: 10.1016/j.radmp.2021.08.004. - DOI
-
- Nilsson R., Liu N.A. Nuclear DNA Damages Generated by Reactive Oxygen Molecules (ROS) Under Oxidative Stress and Their Relevance to Human Cancers, including Ionizing Radiation-Induced Neoplasia Part I: Physical, Chemical and Molecular Biology Aspects. Radiat. Med. Prot. 2020;1:140–152. doi: 10.1016/j.radmp.2020.09.002. - DOI
LinkOut - more resources
Full Text Sources

