A Clinical Validation of a Diagnostic Test for Esophageal Adenocarcinoma Based on a Novel Serum Glycoprotein Biomarker Panel: PromarkerEso
- PMID: 40559996
- PMCID: PMC12196998
- DOI: 10.3390/proteomes13020023
A Clinical Validation of a Diagnostic Test for Esophageal Adenocarcinoma Based on a Novel Serum Glycoprotein Biomarker Panel: PromarkerEso
Abstract
Background: Esophageal adenocarcinoma (EAC) diagnosis involves invasive and expensive endoscopy with biopsy, but rising EAC incidence has not been reduced by increased surveillance. This study aimed to develop and clinically validate a novel glycoprotein biomarker blood test for EAC, named PromarkerEso.
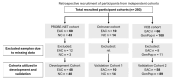
Methods: Serum glycoprotein relative concentrations were measured using a lectin-based magnetic bead array pulldown method, with multiple reaction monitoring mass spectrometry in 259 samples across three independent cohorts. A panel of glycoproteins: alpha-1-antitrypsin, alpha-1-antichymotrypsin, complement C9 and plasma kallikrein, were combined with clinical factors (age, sex and BMI) in an algorithm to categorize the samples by the risk of EAC.
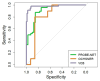
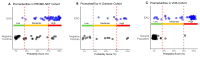
Results: PromarkerEso demonstrated a strong discrimination of EAC from the controls (area under the curve (AUC) of 0.91 in the development cohort and 0.82 and 0.98 in the validation cohorts). The test exhibited a high sensitivity for EAC (98% in the development cohort, and 99.9% and 91% in the validation cohorts) and a high specificity (88% in the development cohort, and 86% and 99% in the validation cohorts). PromarkerEso identified individuals with and without EAC (96% and 95% positive and negative predictive values).
Conclusions: This less invasive approach for EAC detection with the novel combination of these glycoprotein biomarkers and clinical factors coalesces in a potential step toward improved diagnosis.
Keywords: Gastrointestinal diseases; LeMBA; MRM-MS; PromarkerEso; biomarkers; cancer diagnosis; esophageal adenocarcinoma (EAC); glycoprotein; targeted mass spectrometry.
Conflict of interest statement
Scott Bringans, Jordana Sheahan, Iris Wang, Peter Galettis, Kirsten Peters, Richard Lipscombe are employees of Proteomics International, with Scott Bringans, Iris Wang, Kirsten Peters and Richard Lipscombe holding shares in the company. Proteomics International is a beneficiary of patent WO2016077881 A1 that relates to biomarkers described in this manuscript. Michelle Hill is currently the Founder and Managing Director of ProSeek Bio Pty Ltd.; however, the work was conducted during her tenure at QIMR Berghofer Medical Research Institute and has no relationship with ProSeek Bio Pty Ltd.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

