Long-range PCRs and next-generation sequencing to detect cytomegalovirus drug resistance-associated mutations
- PMID: 40560095
- PMCID: PMC12326992
- DOI: 10.1128/aac.00141-25
Long-range PCRs and next-generation sequencing to detect cytomegalovirus drug resistance-associated mutations
Abstract
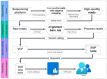
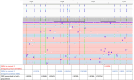
Cytomegalovirus (CMV) infections occur in 10%-40% of organ or stem cell transplant patients. Despite the low prevalence, CMV antiviral resistance has an important impact on patient outcomes. Guidelines for transplant recipients recommend that resistance should be suspected in cases of unchanged or increasing CMV viral loads after a minimum of 2 weeks of antiviral therapy at an appropriate dose or >6 weeks of ganciclovir exposure. Next-generation amplicon sequencing (NGS) makes it possible to directly target the genes involved in this resistance. Currently, six drugs are available, and six CMV genes (UL54-UL97-UL89-UL56-UL51-UL27 genes) can harbor mutations affecting drug efficacy. Here, we developed different primers targeting these six genes with long-range polymerase chain reaction (PCR). Based on clinical requirements, all genes or a subset could be sequenced in a single run using Oxford Nanopore technology and combined with an automatic bioinformatics pipeline to detect and report mutations. We utilized 46 blood samples, five external quality controls, and 10 mixes of two bacmids provided by the national reference center (CNR) Herpesvirus Limoges, each carrying distinct mutations. Assay performance (sensitivity, specificity, and accuracy) was evaluated through an interlaboratory exchange with CNR Herpesvirus. Long-range PCR combined with next-generation sequencing analysis enables earlier and more comprehensive discrimination of the double population and determines whether the detected single-nucleotide polymorphisms are present on single or multiple CMV strains. We developed a next-generation sequencing assay combined with eight long-range PCRs to sequence all genes involved in CMV antiviral resistance and to detect early low-frequency mutations.
Keywords: antiviral resistance; cytomegalovirus; long-range polymerase chain reaction; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Silva Junior HT, Tokat Y, Cai J, Singh I, Sandhu A, Demuth D, Kim J. 2023. Epidemiology, management, and burden of cytomegalovirus in solid organ transplant recipients in selected countries outside of Europe and North America: a systematic review. Transplant Infectious Dis 25:e14070. doi: 10.1111/tid.14070 - DOI - PubMed
-
- Ljungman P, Boeckh M, Hirsch HH, Josephson F, Lundgren J, Nichols G, Pikis A, Razonable RR, Miller V, Griffiths PD, Disease Definitions Working Group of the Cytomegalovirus Drug Development Forum . 2017. Definitions of cytomegalovirus infection and disease in transplant patients for use in clinical trials. Clin Infect Dis 64:87–91. doi: 10.1093/cid/ciw668 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

