High-content image-based pooled screens reveal regulators of synaptogenesis
- PMID: 40560725
- PMCID: PMC12427603
- DOI: 10.1016/j.celrep.2025.115889
High-content image-based pooled screens reveal regulators of synaptogenesis
Abstract
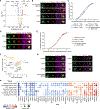
Synapse formation is a fundamental process that shapes the connectivity and function of the nervous system, but the mechanisms regulating synaptogenesis are incompletely understood. Moreover, the interplay of these mechanisms at distinct synapse types remains to be defined. Using a scalable optical pooled screening platform, we investigated the process of synapse induction to uncover modulators of a prototypical synapse-organizing adhesion molecule, neuroligin-1. Analysis of over two million single-cell phenotypic profiles identified 102 candidate regulators of neuroligin-1 that are linked to cell adhesion, cytoskeletal dynamics, and signaling. Among these, we show that the phosphatase PTEN and the dystrophin-associated glycoprotein DAG1 promote neuroligin's roles in inducing presynaptic assembly, with DAG1 selectively regulating inhibitory synapses. This work establishes a scalable high-content screening approach for cell-cell interactions that enables systematic studies of the molecular interactions guiding synaptogenesis.
Keywords: CP: Neuroscience; CRISPR/Cas9; cell adhesion molecules; cell non-autonomous; cell-cell interactions; functional genomics; high-content screening; neuroligin; optical pooled screening; synapse; synaptogenesis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.L. is a consultant to Bifrost Biosystems. P.C.B. is a consultant to and/or holds equity in companies that develop or apply genomic or genome editing technologies: 10x Genomics, General Automation Lab Technologies/Isolation Bio, Next Gen Diagnostics LLC, Cache DNA, Concerto Biosciences, Stately Bio, Ramona Optics, Bifrost Biosystems, and Amber Bio. P.C.B.’s laboratory has received research funding from Calico Life Sciences, Merck, and Genentech for work related to genetic screening.
Figures


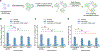


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

