De novo assembly of RNA m6A modification factors into viral genome-associated nuclear bodies drives HCMV RNA accumulation
- PMID: 40560728
- PMCID: PMC12462667
- DOI: 10.1016/j.celrep.2025.115826
De novo assembly of RNA m6A modification factors into viral genome-associated nuclear bodies drives HCMV RNA accumulation
Abstract
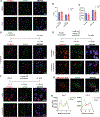
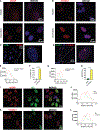
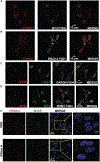
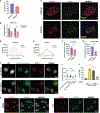
The factors that install and recognize N6-methyladenosine (m6A) on RNA to regulate gene expression are well characterized, but how their spatial organization responds to physiological stress, including infection, is unclear. Here, we show that human cytomegalovirus (HCMV) infection induces accumulation of m6A methyltransferase subunits, including WTAP, together with nuclear m6A reader YTHDC1, into distinctive, membraneless nuclear bodies (NBs) overlapping with incoming virus genomes and immediate-early (IE) RNA transcripts. De novo assembly and integrity of these DNA-associated, IE, virus-activated NBs requires RNAPII transcription, METTL3 m6A methyltransferase activity, and m6A recognition by YTHDC1, but not new protein synthesis. Depleting YTHDC1 or WTAP limits the accumulation of critical HCMV RNAs required for virus DNA replication, interfering with virus reproduction. This reveals a surprising strategy whereby a discrete sub-nuclear RNA biogenesis compartment replete with RNAPII and m6A modification components is swiftly consolidated in proximity to infecting HCMV genomes to initialize and sustain virus gene expression.
Keywords: CP: Microbiology; CP: Molecular biology; RNA m(6)A modification; epitranscriptomics; herpesvirus; infectious stress; nuclear bodies.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE, and Darnell RB (2017). m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006. 10.1101/gad.301036.117. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

