Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization
- PMID: 40560930
- PMCID: PMC12192291
- DOI: 10.1371/journal.pbio.3003223
Ancestral [Fe-S] biogenesis system SMS has a unique mechanism of cluster assembly and sulfur utilization
Abstract
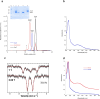
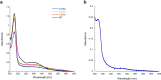
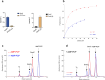
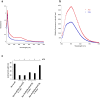
[Fe-S] clusters are ancient and ubiquitous protein co-factors, which contributed to the emergence of life in an anoxic planet. We have recently identified two minimal [Fe-S] biogenesis systems, MIS and SMS, inferred to be ancestral systems dating back to the Last Universal Common Ancestor and which gave rise to the well-studied modern Iron-Sulfur Cluster (ISC), Nitrogen Fixation (NIF), and Sulfur Mobilization (SUF) machineries. The present study focuses on the ancestor SMS from the hyperthermophilic archaeon Methanocaldococcus jannaschii. Biochemical and structural studies showed that SMS is made of a SmsC2B2 heterotetratmer wherein the SmsC subunit hosts both ATP and [Fe-S] cluster binding sites. Binding of ATP and assembly of [Fe-S] were found to be mutually exclusive allowing for a regulatory coupling between binding of both substrates. Mutagenesis and in vitro transfer experiments revealed the key role of SmsC-contained Cys residues in cluster assembly. Strikingly, the SMS system rescued a non-viable Escherichia coli strain lacking endogenous ISC and SUF systems grown under anoxic conditions, in the presence of Na2S, indicating that sulfide is a source of sulfur for SMS. In addition, we predict that most archaea SmsC proteins hold a similar C-terminal [Fe-S] cluster assembly site. Taking into account those unique structural and functional features, we propose a mechanistic model describing how SmsC2B2 assembles and distributes [4Fe-4S] clusters. Altogether this study established SMS as a new bona fide [Fe-S] biogenesis system that operated in anaerobic prokaryotes prior to evolve to SUF after the Great Oxydation Event.
Copyright: © 2025 Dussouchaud et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Fontecave M. Iron–sulfur clusters: ever-expanding roles. Nat Chem Biol. 2006;2:171–4. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

