Cardiomyocyte-derived YOD1 promotes pathological cardiac hypertrophy by deubiquitinating and stabilizing STAT3
- PMID: 40561034
- PMCID: PMC12189954
- DOI: 10.1126/sciadv.adu8422
Cardiomyocyte-derived YOD1 promotes pathological cardiac hypertrophy by deubiquitinating and stabilizing STAT3
Abstract
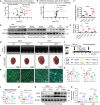
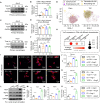
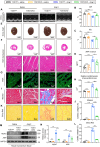
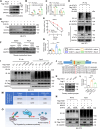
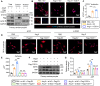
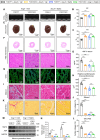
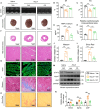
Identifying previously unknown targets for pathological cardiac hypertrophy and understanding its mechanisms are crucial. Here, we observed that the deubiquitinating enzyme YOD1 was moderately elevated in human hypertrophic myocardium and mouse models. Cardiomyocyte-specific knockout of YOD1 reduced Ang II- and TAC-induced cardiac hypertrophy. Subsequently, we used multiple proteomic analyses to identify and confirm STAT3 as a substrate protein for YOD1. Mechanistically, our findings revealed that the C155 site of YOD1 removes K48-linked ubiquitin chains from K97 on STAT3, stabilizing STAT3 levels and enhancing its nuclear translocation in cardiomyocytes under Ang II stimulation. Notably, inhibiting STAT3 reversed the antihypertrophic effects of YOD1 deficiency in Ang II-challenged mice. In addition, pharmacological inhibition of YOD1 mitigated Ang II-induced pathological ventricular remodeling in mice. This study clarifies the role of YOD1 and introduces a previously unidentified YOD1-STAT3 axis in regulating pathological cardiac hypertrophy, providing valuable insights for drug development targeting this condition.
Figures

References
-
- Messerli F. H., Williams B., Ritz E., Essential hypertension. Lancet 370, 591–603 (2007). - PubMed
-
- Oldfield C. J., Duhamel T. A., Dhalla N. S., Mechanisms for the transition from physiological to pathological cardiac hypertrophy. Can. J. Physiol. Pharmacol. 98, 74–84 (2020). - PubMed
-
- McKinsey T. A., Kass D. A., Small-molecule therapies for cardiac hypertrophy: Moving beneath the cell surface. Nat. Rev. Drug Discov. 6, 617–635 (2007). - PubMed
-
- Wang B., Cai W., Ai D., Zhang X., Yao L., The role of deubiquitinases in vascular diseases. J. Cardiovasc. Transl. Res. 13, 131–141 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

