Impact of the tumor immune contexture in microsatellite-stable metastatic colorectal cancer treated with avelumab, cetuximab, and irinotecan
- PMID: 40562041
- PMCID: PMC12281371
- DOI: 10.1016/j.xcrm.2025.102201
Impact of the tumor immune contexture in microsatellite-stable metastatic colorectal cancer treated with avelumab, cetuximab, and irinotecan
Abstract
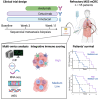
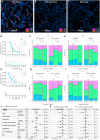
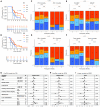
The treatment of patients with microsatellite-stable (MSS) metastatic colorectal cancer (mCRC) remains a significant clinical challenge. Cetuximab, an anti-epidermal growth factor receptor (EGFR) monoclonal antibody (mAb), induces immunogenic cell death, potentially synergizing with immune checkpoint inhibitors. The phase 2, proof-of-concept, single-arm AVETUXIRI trial (ClinicalTrials.gov: NCT03608046) evaluates the safety and efficacy of cetuximab, irinotecan (a topoisomerase I inhibitor), and avelumab (an anti-programmed cell death ligand 1 [PD-L1]) in 57 patients with RAS wild-type or mutated MSS mCRC refractory to chemotherapy and anti-EGFR mAbs. Exploratory objectives include investigating the tumor immune microenvironment within mCRC biopsies performed during the trial and correlating it with treatment activity. A manageable safety profile is observed. Although the overall efficacy endpoints are not met, biomarkers associated with clinical efficacy are identified. Patients exhibiting a high Immunoscore, strong cytotoxic and T cell proximity to tumor cells, and a high genetic immunoediting score within mCRC biopsies before treatment demonstrate significant therapeutic survival benefit, independent of RAS tumor mutation status.
Keywords: Immunoscore; RAS mutation; avelumab; biomarkers; cetuximab; immunoediting score; immunofluorescence; immunotherapy; metastatic colorectal cancer; transcriptomics.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflicts of interest.
Figures



References
-
- Cervantes A., Adam R., Roselló S., Arnold D., Normanno N., Taïeb J., Seligmann J., De Baere T., Osterlund P., Yoshino T., et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023;34:10–32. doi: 10.1016/j.annonc.2022.10.003. - DOI - PubMed
-
- Arnold D., Lueza B., Douillard J.Y., Peeters M., Lenz H.J., Venook A., Heinemann V., Van Cutsem E., Pignon J.P., Tabernero J., et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann. Oncol. 2017;28:1713–1729. doi: 10.1093/annonc/mdx175. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

