PADI4-mediated citrullination of histone H3 stimulates HIV-1 transcription
- PMID: 40562767
- PMCID: PMC12198384
- DOI: 10.1038/s41467-025-61029-0
PADI4-mediated citrullination of histone H3 stimulates HIV-1 transcription
Abstract
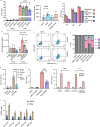
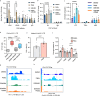
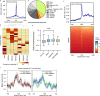
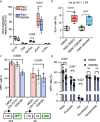
HIV-1 infection establishes a reservoir of long-lived cells with integrated proviral DNA that can persist despite antiretroviral therapy (ART). Certain reservoir cells can be reactivated to reinitiate infection. The mechanisms governing proviral latency and transcriptional regulation of the provirus are complex. Here, we identify a role for histone H3 citrullination, a post-translational modification catalyzed by protein-arginine deiminase type-4 (PADI4), in HIV-1 transcription and latency. PADI4 inhibition by the small molecule inhibitor GSK484 reduces HIV-1 transcription after T cell activation in ex vivo cultures of CD4+ T cells from people living with HIV-1 in a cross-sectional study. The effect is more pronounced in individuals with active viremia compared to individuals under effective ART. Cell models of HIV-1 latency show that citrullination of histone H3 occurs at the HIV-1 promoter upon T cell stimulation, which facilitates proviral transcription. HIV-1 integrates into genomic regions marked by H3 citrullination and these proviruses are less prone to latency compared to those in non-citrullinated chromatin. Inhibiting PADI4 leads to compaction of the HIV-1 promoter chromatin and an increase of heterochromatin protein 1α (HP1α)-covered heterochromatin, in a mechanism partly dependent on the HUSH complex. Our data reveal a novel mechanism to explain HIV-1 latency and transcriptional regulation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

