Developing drug-like single-domain antibodies (VHH) from in vitro libraries
- PMID: 40562779
- PMCID: PMC12203854
- DOI: 10.1080/19420862.2025.2516676
Developing drug-like single-domain antibodies (VHH) from in vitro libraries
Abstract
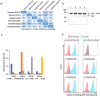
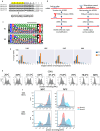
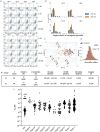
Here, we describe a new VHH library for therapeutic discovery which optimizes humanness, stability, affinity, diversity, developability, and facile purification using protein A in the absence of an Fc domain. Four therapeutic humanized VHHs were used as scaffolds, into which we inserted human HCDR1s, HCDR2s and HCDR3s. The HCDR1 and HCDR2 sequences were derived from human VH3 family next-generation sequencing datasets informatically purged of sequence liabilities, synthesized as array-based oligonucleotides, cloned as single CDR libraries into each of the parental scaffolds and filtered for protein A binding by yeast display to ensure correct folding and display. After filtering, the CDR1 and CDR2 libraries were combined with amplified human HCDR3 from human CD19+ IgM+ B cells. This library was further improved by eliminating long consecutive stretches of tyrosines in CDR3 and enriching for CDR1-2 diversity with elevated tolerance to high temperatures. A broad diversity of high affinity (100 pM-10 nM), developable binders was directly isolated, with developability evaluated for most assays using the isolated VHHs, rather than fused to Fc, which is customary. This represents the first systematic developability assessment of isolated VHH molecules.
Keywords: Antibody; VHH; camelid; drug discovery; phage display; single domain; yeast display.
Conflict of interest statement
MFE, EM, LARC, DK, HT, JL, RDN, AF, AD, LS, SD, FF, ARMB are or were employees of Specifica, an IQVIA business at the time of writing. CLL is currently an employee of Sanofi, Cambidge, USA. All other authors declare no conflicts of interest. AART and ARMB are listed as co-inventors on patent describing this technology.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
