Maturation of human cardiac organoids enables complex disease modeling and drug discovery
- PMID: 40562874
- PMCID: PMC12259470
- DOI: 10.1038/s44161-025-00669-3
Maturation of human cardiac organoids enables complex disease modeling and drug discovery
Abstract
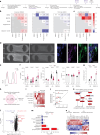
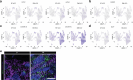
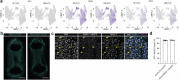
Maturation of human pluripotent stem (hPS) cell-derived cardiomyocytes is critical for their use as a model system. Here we mimic human heart maturation pathways in the setting of hPS cell-derived cardiac organoids (hCOs). Specifically, transient activation of 5' AMP-activated protein kinase and estrogen-related receptor enhanced cardiomyocyte maturation, inducing expression of mature sarcomeric and oxidative phosphorylation proteins, and increasing metabolic capacity. hCOs generated using the directed maturation protocol (DM-hCOs) recapitulate cardiac drug responses and, when derived from calsequestrin 2 (CASQ2) and ryanodine receptor 2 (RYR2) mutant hPS cells exhibit a pro-arrhythmia phenotype. These DM-hCOs also comprise multiple cell types, which we characterize and benchmark to the human heart. Modeling of cardiomyopathy caused by a desmoplakin (DSP) mutation resulted in fibrosis and cardiac dysfunction and led to identifying the bromodomain and extra-terminal inhibitor INCB054329 as a drug mitigating the desmoplakin-related functional defect. These findings establish DM-hCOs as a versatile platform for applications in cardiac biology, disease and drug screening.
© 2025. Crown.
Conflict of interest statement
Competing interests: E.R.P., R.J.M. and J.E.H. are co-inventors on a patent relating to the Heart-Dyno device and human cardiac organoid maturation used in this study (WO2018035574A1 filed by the University of Queensland) which is licensed to Dynomics. E.R.P., R.J.M. and J.E.H. are co-inventors on a patent for cardiac regeneration therapeutics (WO2020186283A1 filed by QIMR Berghofer). E.R.P. and J.E.H. are co-inventors on a patent for the serum-free conditions supporting the vascular population used in this study (WO2024016058 filed by MCRI and QIMR Berghofer). J.E.H. is co-inventor on licensed patents for cardiac differentiation and engineered heart muscle, some aspects of which are used in this study (WO2015040142A1 and WO2015025030A1), which are licensed to MyriaMed and Repairon. E.R.P., R.J.M. and J.E.H. are cofounders, scientific advisors, and stockholders in Dynomics. J.E.H. and R.J.M. are co-inventors on a provisional patent filed by QIMR Berghofer on the DM-hCO conditions discovered in this study (2024902826). The remaining authors declare no competing interests.
Figures






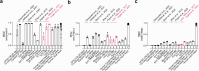





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
