AIMP3 maintains cardiac homeostasis by regulating the editing activity of methionyl-tRNA synthetase
- PMID: 40562875
- PMCID: PMC12259467
- DOI: 10.1038/s44161-025-00670-w
AIMP3 maintains cardiac homeostasis by regulating the editing activity of methionyl-tRNA synthetase
Abstract
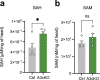
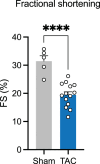
In mammals, nine aminoacyl tRNA synthetases (ARSs) and three auxiliary proteins (ARS-interacting multifunctional proteins 1-3 (AIMP1-3)) form the multisynthetase complex (MSC), a molecular hub that provides a subset of aminoacylated tRNAs to the ribosome and partakes in translation-independent signaling. Knowledge of the role of AIMPs in organ physiology is currently limited. AIMP3 (also known as EEF1E1) was proposed to anchor methionyl tRNA synthetase (MetRS) in the complex and regulate protein synthesis through translation initiation and elongation. Here we show that a cardiomyocyte-specific conditional knockout of AIMP3 in mice leads to lethal cardiomyopathy. MetRS localization, aminoacylation efficiency and global protein synthesis were unaffected in our model, suggesting an alternative mechanism for the pathology. We found that AIMP3 is essential for homocysteine editing by MetRS, a reaction that is necessary for the maintenance of translation fidelity. Homocysteine accumulation induced reactive oxygen species production, protein aggregation, mitochondrial dysfunction, autophagy and ultimately cell death.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






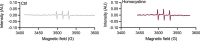
Similar articles
-
Ring Finger Protein 2 Promotes Oxidative Stress and Mitochondrial Dysfunction in Doxorubicin-Induced Cardiotoxicity Via the Mercaptopyruvate Sulfurtransferase/Hydrogen Sulfide Pathway.J Am Heart Assoc. 2025 Aug 19;14(16):e041440. doi: 10.1161/JAHA.125.041440. Epub 2025 Aug 6. J Am Heart Assoc. 2025. PMID: 40767300
-
AIMP3/p18 controls translational initiation by mediating the delivery of charged initiator tRNA to initiation complex.J Mol Biol. 2012 Nov 2;423(4):475-81. doi: 10.1016/j.jmb.2012.07.020. Epub 2012 Aug 4. J Mol Biol. 2012. PMID: 22867704
-
Acid sphingomyelinase promotes diabetic cardiomyopathy via disruption of mitochondrial calcium homeostasis.Cardiovasc Diabetol. 2025 Jul 10;24(1):272. doi: 10.1186/s12933-025-02801-w. Cardiovasc Diabetol. 2025. PMID: 40640752 Free PMC article.
-
Intensive case management for severe mental illness.Cochrane Database Syst Rev. 2010 Oct 6;(10):CD007906. doi: 10.1002/14651858.CD007906.pub2. Cochrane Database Syst Rev. 2010. Update in: Cochrane Database Syst Rev. 2017 Jan 06;1:CD007906. doi: 10.1002/14651858.CD007906.pub3. PMID: 20927766 Free PMC article. Updated.
-
From gene to mechanics: a comprehensive insight into the mechanobiology of LMNA mutations in cardiomyopathy.Cell Commun Signal. 2024 Mar 27;22(1):197. doi: 10.1186/s12964-024-01546-5. Cell Commun Signal. 2024. PMID: 38539233 Free PMC article. Review.
References
-
- Jakubowski, H. Quality control in tRNA charging. Wiley Interdiscip. Rev. RNA3, 295–310 (2012). - PubMed
MeSH terms
Substances
Grants and funding
- S10 OD023461/OD/NIH HHS/United States
- HL154001/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 AG079842/AG/NIA NIH HHS/United States
- HL136951/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- S10 OD025008/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
