Fine-mapping genomic loci refines bipolar disorder risk genes
- PMID: 40562893
- PMCID: PMC12229890
- DOI: 10.1038/s41593-025-01998-z
Fine-mapping genomic loci refines bipolar disorder risk genes
Erratum in
-
Author Correction: Fine-mapping genomic loci refines bipolar disorder risk genes.Nat Neurosci. 2025 Oct 27. doi: 10.1038/s41593-025-02133-8. Online ahead of print. Nat Neurosci. 2025. PMID: 41145888 No abstract available.
Abstract
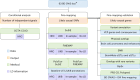
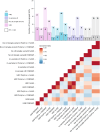
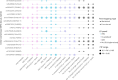
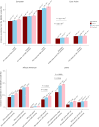
Bipolar disorder is a heritable mental illness with complex etiology. While the largest published genome-wide association study identified 64 bipolar disorder risk loci, the causal SNPs and genes within these loci remain unknown. We applied a suite of statistical and functional fine-mapping methods to these loci and prioritized 17 likely causal SNPs for bipolar disorder. We mapped these SNPs to genes and investigated their likely functional consequences by integrating variant annotations, brain cell-type epigenomic annotations, brain quantitative trait loci and results from rare variant exome sequencing in bipolar disorder. Convergent lines of evidence supported the roles of genes involved in neurotransmission and neurodevelopment, including SCN2A, TRANK1, DCLK3, INSYN2B, SYNE1, THSD7A, CACNA1B, TUBBP5, FKBP2, RASGRP1, FURIN, FES, MED24 and THRA among others in bipolar disorder. These represent promising candidates for functional experiments to understand biological mechanisms and therapeutic potential. Additionally, we demonstrated that fine-mapping effect sizes can improve performance of bipolar disorder polygenic risk scores across diverse populations and present a high-throughput fine-mapping pipeline.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: O.A.A. has served as a speaker for Janssen, Lundbeck and Sunovion, and as a consultant for Cortechs.ai. S.K.S. has served as speaker for Janssen, Takeda and Medice Arzneimittel Puetter GmbH & CoKG. E.V. has received grants and served as consultant, advisor or CME speaker for the following entities (unrelated to the present work): AB-Biotics, Abbott, AbbVie, Adamed, Angelini, Biogen, Biohaven, Boehringer Ingelheim, Casen-Recordati, Celon, Compass, Dainippon Sumitomo Pharma, Ethypharm, Ferrer, Gedeon Richter, GH Research, Glaxo Smith-Kline, Idorsia, Janssen, Johnson & Johnson, Lundbeck, Newron, Novartis, Organon, Otsuka, Rovi, Sage, Sanofi-Aventis, Sunovion, Takeda and Viatris. P.B.M. has received remuneration from Janssen (Australia) and Sanofi (Hangzhou) for lectures, and Janssen (Australia) for advisory board membership. M.O.D. and M.J.O. have received grants from Akrivia Health and Takeda Pharmaceuticals for work unrelated to this project. The remaining authors declare no competing interests.
Figures

Update of
-
Fine-mapping genomic loci refines bipolar disorder risk genes.medRxiv [Preprint]. 2024 Sep 17:2024.02.12.24302716. doi: 10.1101/2024.02.12.24302716. medRxiv. 2024. Update in: Nat Neurosci. 2025 Jul;28(7):1393-1403. doi: 10.1038/s41593-025-01998-z. PMID: 38405768 Free PMC article. Updated. Preprint.
References
-
- Craddock, N. & Sklar, P. Genetics of bipolar disorder. Lancet381, 1654–1662 (2013). - PubMed
-
- McGuffin, P. et al. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry60, 497–502 (2003). - PubMed
-
- Smoller, J. W. & Finn, C. T. Family, twin, and adoption studies of bipolar disorder. Am. J. Med. Genet. C Semin. Med. Genet.123C, 48–58 (2003). - PubMed

