Separation-of-Function Alleles of smc-5 Reveal Domain-Specific Defects and a Conserved Residue Critical for Genome Maintenance
- PMID: 40563397
- PMCID: PMC12191127
- DOI: 10.3390/biom15060755
Separation-of-Function Alleles of smc-5 Reveal Domain-Specific Defects and a Conserved Residue Critical for Genome Maintenance
Abstract
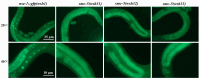
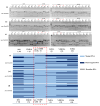
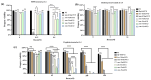
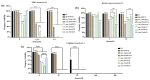
The SMC-5/6 complex safeguards genome stability through the coordinated action of its core SMC proteins and associated NSE subunits. NSE-1 is a key component of the complex and is essential for DNA repair, yet it remains poorly characterized in Caenorhabditis elegans. To further elucidate the functional mechanisms of NSE-1, we performed an EMS-based forward genetic screen in an nse-1::gfp(wsh1) reporter strain to identify mutants with defective NSE-1 expression or nuclear localization. We isolated three mutants; smc-5(wsh31), smc-5(wsh32), and smc-5(wsh33), that display impaired NSE-1::GFP nuclear localization. SNP mapping and whole-genome sequencing revealed three novel smc-5 alleles: two truncations, alleles smc-5(wsh31) (C587*) and smc-5(wsh32) (Q655*), and one missense variant, smc-5(wsh33) (Y975D), each altering a highly conserved residue in the SMC domain. All three mutants exhibited significantly reduced brood size, progeny viability, and slightly elevated male percentages. Phenotypic characterization revealed that the truncations completely abrogate NSE-1::GFP nuclear localization, whereas the missense allele causes stage-dependent, partial mislocalization. Functional assays further demonstrated allele-specific and developmental stage-dependent hypersensitivities to DNA-damaging agents (MMS, HU, and cisplatin). These separation-of-function smc-5 alleles underscore the importance of domains and conserved residues in complex integrity and genome maintenance, and provide powerful genetic tools to dissect SMC-5/6 functions in vivo.
Keywords: Caenorhabditis elegans; NSE-1 localization; SMC-5/6 complex; genome stability; smc-5.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Mutant alleles of the Caenorhabditis elegans rde-1 gene identified through chemical mutagenesis of an snRNA misprocessing reporter.G3 (Bethesda). 2025 Jul 9;15(7):jkaf097. doi: 10.1093/g3journal/jkaf097. G3 (Bethesda). 2025. PMID: 40324366 Free PMC article.
-
Caenorhabditis elegans NSE3 homolog (MAGE-1) is involved in genome stability and acts in inter-sister recombination during meiosis.Genetics. 2023 Oct 4;225(2):iyad149. doi: 10.1093/genetics/iyad149. Genetics. 2023. PMID: 37579186 Free PMC article.
-
Interdependence of Pasha and Drosha for localization and function of the Microprocessor in C. elegans.Nat Commun. 2025 Jul 1;16(1):5595. doi: 10.1038/s41467-025-60721-5. Nat Commun. 2025. PMID: 40595566 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

