Gut Microbiota and Neurovascular Patterns in Amnestic Mild Cognitive Impairment
- PMID: 40563710
- PMCID: PMC12190368
- DOI: 10.3390/brainsci15060538
Gut Microbiota and Neurovascular Patterns in Amnestic Mild Cognitive Impairment
Abstract
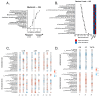
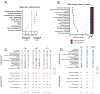
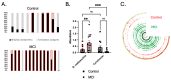
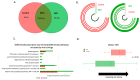
Background/Objectives: The interplay between the gut microbiome (GMB) and neurovascular function in neurodegeneration is unclear. The goal of this proof-of-concept, cross-sectional study is to identify relationships between the GMB, neurovascular functioning, and cognition in amnestic mild cognitive impairment (aMCI), the prototypical prodromal symptomatic stage of Alzheimer's disease (AD). Methods: Participants (n = 14 aMCI and 10 controls) provided fecal samples for GMB sequencing (16S and shotgun metagenomics), underwent MRI, and completed cognitive testing. Cerebral vascular reactivity (CVR), cerebral blood flow (CBF), and arterial transit time (ATT) were assessed. Statistical analyses evaluated the relationships between discriminatory taxa, cerebrovascular metrics, and cognition. Results: Sequencing revealed differentially abundant bacterial and viral taxa distinguishing aMCI from controls. Spearman correlations revealed that bacteria known to induce inflammation were negatively associated with CVR, CBF, and cognition, and positively associated with ATT. A reciprocal pattern emerged for the association of taxa with gut health. Conclusions: Our results provide preliminary evidence that pro-inflammatory gut bacterial and viral taxa are associated with neurovascular dysfunction and cognitive impairment in prodromal AD, highlighting their potential as candidate microbial biomarkers and targets for early intervention.
Keywords: Alzheimer’s disease; blood–brain barrier; dementia; microbiome; neurodegeneration; neurovascular dysfunction; shotgun metagenomics; virome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Cerebral perfusion correlates with amyloid deposition in patients with mild cognitive impairment due to Alzheimer's disease.J Prev Alzheimers Dis. 2025 Feb;12(2):100031. doi: 10.1016/j.tjpad.2024.100031. Epub 2025 Jan 1. J Prev Alzheimers Dis. 2025. PMID: 39863326 Free PMC article.
-
Donepezil for dementia due to Alzheimer's disease.Cochrane Database Syst Rev. 2018 Jun 18;6(6):CD001190. doi: 10.1002/14651858.CD001190.pub3. Cochrane Database Syst Rev. 2018. PMID: 29923184 Free PMC article.
-
CSF and blood biomarkers for the diagnosis of Alzheimer's disease: a systematic review and meta-analysis.Lancet Neurol. 2016 Jun;15(7):673-684. doi: 10.1016/S1474-4422(16)00070-3. Epub 2016 Apr 8. Lancet Neurol. 2016. PMID: 27068280
-
CSF tau and the CSF tau/ABeta ratio for the diagnosis of Alzheimer's disease dementia and other dementias in people with mild cognitive impairment (MCI).Cochrane Database Syst Rev. 2017 Mar 22;3(3):CD010803. doi: 10.1002/14651858.CD010803.pub2. Cochrane Database Syst Rev. 2017. PMID: 28328043 Free PMC article.
-
Cholinesterase inhibitors for vascular dementia and other vascular cognitive impairments: a network meta-analysis.Cochrane Database Syst Rev. 2021 Feb 22;2(2):CD013306. doi: 10.1002/14651858.CD013306.pub2. Cochrane Database Syst Rev. 2021. PMID: 33704781 Free PMC article.
References
-
- Jack C.R., Jr., Andrews J.S., Beach T.G., Buracchio T., Dunn B., Graf A., Hansson O., Ho C., Jagust W., McDade E., et al. Revised criteria for diagnosis and staging of Alzheimer’s disease: Alzheimer’s Association Workgroup. Alzheimers Dement. 2024;20:5143–5169. doi: 10.1002/alz.13859. - DOI - PMC - PubMed
-
- Jack C.R., Jr., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., Holtzman D.M., Jagust W., Jessen F., Karlawish J., et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. - DOI - PMC - PubMed
-
- Ferreiro A.L., Choi J., Ryou J., Newcomer E.P., Thompson R., Bollinger R.M., Hall-Moore C., Ndao I.M., Sax L., Benzinger T.L.S., et al. Gut microbiome composition may be an indicator of preclinical Alzheimer’s disease. Sci. Transl. Med. 2023;15:eabo2984. doi: 10.1126/scitranslmed.abo2984. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

