Orchestration of Gut-Liver-Associated Transcription Factors in MAFLD: From Cross-Organ Interactions to Therapeutic Innovation
- PMID: 40564141
- PMCID: PMC12191254
- DOI: 10.3390/biomedicines13061422
Orchestration of Gut-Liver-Associated Transcription Factors in MAFLD: From Cross-Organ Interactions to Therapeutic Innovation
Abstract
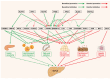
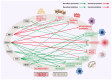
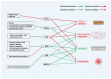
Metabolic dysfunction-associated fatty liver disease (MAFLD) represents a global health burden, however, therapeutic advancements remain hindered by incomplete insights on mechanisms and suboptimal clinical interventions. This review focused on the transcription factors (TFs) associated with the gut-liver axis, emphasizing their roles as molecular interpreters of systemic crosstalk in MAFLD. We delineate how TF networks integrate metabolic, immune, and gut microbial signals to manage hepatic steatosis, inflammation, and fibrosis. For instance, metabolic TFs such as peroxisome proliferator-activated receptor α (PPARα) and farnesoid X receptor (FXR) are responsible for regulating lipid oxidation and bile acid homeostasis, while immune-related TFs like signal transducer and activator of transcription 3 (STAT3) modulate inflammatory cascades involving immune cells. Emerging evidence highlights microbiota-responsive TFs, like hypoxia-inducible factor 2α (HIF2α) and aryl hydrocarbon receptor (AHR), linking microbial metabolite signaling to hepatic metabolic reprogramming. Critically, TF-centric therapeutic strategies, including selective TF-agonists, small molecules targeted to degrade TF, and microbiota modulation, hold considerable promise for treating MAFLD. By synthesizing these insights, this review underscores the necessity to dissect TF-mediated interorgan communication and proposes a roadmap for translating mechanism discoveries into precision therapies. Future research should prioritize the use of multi-omics approaches to map TF interactions and validate their clinical relevance to MAFLD.
Keywords: inflammation; metabolic dysfunction-associated fatty liver disease; metabolism; microbiota; molecular mechanisms; transcription factors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

