Applications of Surface Plasmon Resonance in Heparan Sulfate Interactome Research
- PMID: 40564190
- PMCID: PMC12191165
- DOI: 10.3390/biomedicines13061471
Applications of Surface Plasmon Resonance in Heparan Sulfate Interactome Research
Abstract
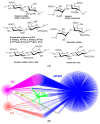
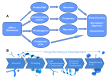
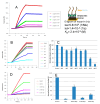
Surface plasmon resonance (SPR) is a powerful tool for analyzing biomolecular interactions and is widely used in basic biomedical research and drug discovery. Heparan sulfate (HS) is a linear complex polysaccharide and a key component of the extracellular matrix and cell surfaces. HS plays a pivotal role in maintaining cellular functions and tissue homeostasis by interacting with numerous proteins, making it essential for normal physiological processes and disease states. Deciphering the interactome of HS unlocks the mechanisms underlying its biological functions and the potential for novel HS-related therapeutics. This review presents an overview of the recent advances in the application of SPR technology to HS interactome research. We discuss methodological developments, emerging trends, and key findings that illustrate how SPR is expanding our knowledge of HS-mediated molecular interactions. Additionally, we highlight the potential of SPR-based approaches in identifying novel therapeutic targets and developing HS-mimetic drugs, thereby opening new avenues for intervention in HS-related diseases.
Keywords: heparan sulfate; heparin; interactome; surface plasmon resonance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
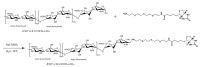


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

