Identification Exploring the Mechanism and Clinical Validation of Mitochondrial Dynamics-Related Genes in Membranous Nephropathy Based on Mendelian Randomization Study and Bioinformatics Analysis
- PMID: 40564208
- PMCID: PMC12191289
- DOI: 10.3390/biomedicines13061489
Identification Exploring the Mechanism and Clinical Validation of Mitochondrial Dynamics-Related Genes in Membranous Nephropathy Based on Mendelian Randomization Study and Bioinformatics Analysis
Abstract
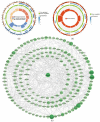
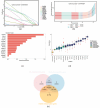
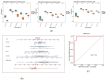
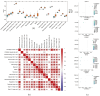
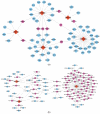
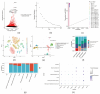
Background: Membranous nephropathy (MN), a prevalent glomerular disorder, remains poorly understood in terms of its association with mitochondrial dynamics (MD). This study investigated the mechanistic involvement of mitochondrial dynamics-related genes (MDGs) in the pathogenesis of MN. Methods: Comprehensive bioinformatics analyses-encompassing Mendelian randomization, machine-learning algorithms, and single-cell RNA sequencing (scRNA-seq)-were employed to interrogate transcriptomic datasets (GSE200828, GSE73953, and GSE241302). Core MDGs were further validated using reverse-transcription quantitative polymerase chain reaction (RT-qPCR). Results: Four key MDGs-RTTN, MYO9A, USP40, and NFKBIZ-emerged as critical determinants, predominantly enriched in olfactory transduction pathways. A nomogram model exhibited exceptional diagnostic performance (area under the curve [AUC] = 1). Seventeen immune cell subsets, including regulatory T cells and activated dendritic cells, demonstrated significant differential infiltration in MN. Regulatory network analyses revealed ATF2 co-regulation mediated by RTTN and MYO9A, along with RTTN-driven modulation of ELOA-AS1 via hsa-mir-431-5p. scRNA-seq analysis identified mesenchymal-epithelial transitioning cells as key contributors, with pseudotime trajectory mapping indicating distinct temporal expression profiles: NFKBIZ (initial upregulation followed by decline), USP40 (gradual fluctuation), and RTTN (persistently low expression). RT-qPCR results corroborated a significant downregulation of all four genes in MN samples compared to controls (p < 0.05). Conclusions: These findings elucidate the molecular underpinnings of MDG-mediated mechanisms in MN, revealing novel diagnostic biomarkers and therapeutic targets. The data underscore the interplay between mitochondrial dynamics and immune dysregulation in MN progression, providing a foundation for precision medicine strategies.
Keywords: machine learning; membranous nephropathy; mendelian randomization; mitochondrial dynamics; single-cell analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




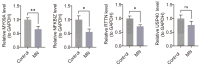
References
-
- Prunotto M., Nachman P.H., Gillespie B.S., Beck L.H., Jr., Thompson A.M., Hu A.H., Stafford E.A., Tarnoff J.M., Rovin B.H. Designing clinical trials for the treatment of membranous nephropathy in the anti-PLA2R era: Results of a Nephcure membranous nephropathy workshop. Kidney Int. 2025;107:809–815. doi: 10.1016/j.kint.2024.09.020. - DOI - PubMed
-
- Storrar J., Gill-Taylor T., Chinnadurai R., Chrysochou C., Poulikakos D., Rainone F., Ritchie J., Lamerton E., Kalra P.A., Sinha S. A low rate of end-stage kidney disease in membranous nephropathy: A single centre study over 2 decades. PLoS ONE. 2022;17:e0276053. doi: 10.1371/journal.pone.0276053. - DOI - PMC - PubMed
-
- Li H., Zhou L., Zhou W., Zhang X., Shang J., Feng X., Yu L., Fan J., Ren J., Zhang R., et al. Decoding the mitochondrial connection: Development and validation of biomarkers for classifying and treating systemic lupus erythematosus through bioinformatics and machine learning. BMC Rheumatol. 2023;7:44. doi: 10.1186/s41927-023-00369-0. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

