Genome-Wide Association Study for Individual Primal Cut Quality Traits in Canadian Commercial Crossbred Pigs
- PMID: 40564306
- PMCID: PMC12189728
- DOI: 10.3390/ani15121754
Genome-Wide Association Study for Individual Primal Cut Quality Traits in Canadian Commercial Crossbred Pigs
Abstract
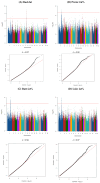
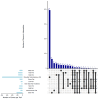
This study identified genomic variants and potential candidate genes associated with 11 primal cut traits (back fat, belly fat, total fat, loin fat, ham fat, picnic fat, butt fat, loin intramuscular fat content, ham side fat, shoulder dorsal fat, and belly side fat thicknesses) in Canadian commercial crossbred pigs. Genome-wide association studies using whole genome sequencing data were conducted using genotyping data from 1118 commercial crossbred pigs. This analysis revealed multiple QTLs across chromosomes SSC1, 2, 3, 6, 7, 9, 14, 15, and 17, associated with fat traits. Notably, an SNP at position 160,230,075 bp on SSC1 was significantly associated with multiple fat traits, including belly fat, butt fat, ham fat, loin fat, picnic fat, and side fat. Common genes in windows associated with multiple traits, such as MC4R, RNF152, and CDH20 were shared across these traits, suggesting pleiotropic effects. Some of the QTLs were near previously identified QTLs or candidate genes that have been reported to be linked to meat quality traits associated with backfat and intramuscular fat. Other candidate genes identified in the study include TNFRSF11A, LEPR, and genes from the SERPINB family, highlighting their roles in fat deposition and composition. Additional candidate genes were also implicated in regulation of fat metabolism, adipogenesis, and adiposity. These findings offer valuable insights into the genetic architecture of fat traits in pigs, which could inform breeding strategies aimed at improving the pork quality.
Keywords: SNPs; fat metabolism; pork quality; primal fat; whole genome sequencing.
Conflict of interest statement
Kerry Houlahan and Robert Kemp were employees of Genesus Genetic Technology Inc. at the time of the study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Identification of new candidate genes affecting drip loss in pigs based on genomics and transcriptomics data.J Anim Sci. 2025 Jan 4;103:skaf177. doi: 10.1093/jas/skaf177. J Anim Sci. 2025. PMID: 40485044 Free PMC article.
-
Genetic parameters for quality attributes in individual pork primal cuts.Meat Sci. 2025 Oct;228:109887. doi: 10.1016/j.meatsci.2025.109887. Epub 2025 Jun 13. Meat Sci. 2025. PMID: 40543385
-
Sequence-based GWAS reveals genes and variants associated with predicted methane emissions in French dairy cows.Genet Sel Evol. 2025 Jun 17;57(1):32. doi: 10.1186/s12711-025-00977-z. Genet Sel Evol. 2025. PMID: 40528185 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Li J., Wu J., Jian Y., Zhuang Z., Qiu Y., Huang R., Lu P., Guan X., Huang X., Li S., et al. Genome-Wide Association Studies Revealed Significant QTLs and Candidate Genes Associated with Backfat and Loin Muscle Area in Pigs Using Imputation-Based Whole Genome Sequencing Data. Animals. 2022;12:2911. doi: 10.3390/ani12212911. - DOI - PMC - PubMed
-
- Miar Y., Plastow G.S., Moore S.S., Manafiazar G., Charagu P., Kemp R.A., van Haandel B., Huisman A.E., Zhang C.Y., Mckay R.M., et al. Genetic and Phenotypic Parameters for Carcass and Meat Quality Traits in Commercial Crossbred Pigs. J. Anim. Sci. 2014;92:2869–2884. doi: 10.2527/jas.2014-7685. - DOI - PubMed
-
- Ciobanu D.C., Lonergan S.M., Huff-Lonergan E.J. The Genetics of the Pig. CABI; Wallingford, UK: 2011. Genetics of Meat Quality and Carcass Traits; pp. 355–389.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

