New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine
- PMID: 40564859
- PMCID: PMC12192280
- DOI: 10.3390/diagnostics15121539
New Generation of Clinical Epigenetics Analysis and Diagnosis for Precision Medicine
Abstract
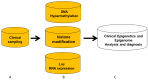
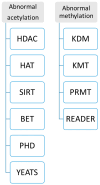
Following the application of epigenetic and epigenomics research into tumor diseases, cardiovascular disease, diabetes, hereditary diseases, and rare diseases, in vitro diagnostics (IVD) epigenetic and epigenomics are increasingly employed for those patients. Here, we review a clinical sampling of epigenetics and epigenomics from patients. We then present procedures, including the detection procedure of clinical epigenetic approaches from clinical samples, clinical epigenomic methods applied to those samples, the small cell number of epigenetics, and epigenomics. Finally, we present the current IVD of epigenetics and epigenomics used for clinical analysis and diagnosis, along with the development of approaches. To improve clinical study and diagnosis, we also introduce clinical research and clinical applications to develop a more comprehensive strategy to maximize the sensitivity and specificity of the epigenetics and epigenomics analysis and diagnosis.
Keywords: epigenetics; epigenomics; in vitro diagnostics (IVD); single-cell diagnosis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Rapid molecular tests for tuberculosis and tuberculosis drug resistance: a qualitative evidence synthesis of recipient and provider views.Cochrane Database Syst Rev. 2022 Apr 26;4(4):CD014877. doi: 10.1002/14651858.CD014877.pub2. Cochrane Database Syst Rev. 2022. PMID: 35470432 Free PMC article.
-
Factors that impact on the use of mechanical ventilation weaning protocols in critically ill adults and children: a qualitative evidence-synthesis.Cochrane Database Syst Rev. 2016 Oct 4;10(10):CD011812. doi: 10.1002/14651858.CD011812.pub2. Cochrane Database Syst Rev. 2016. PMID: 27699783 Free PMC article.
-
Magnetic resonance perfusion for differentiating low-grade from high-grade gliomas at first presentation.Cochrane Database Syst Rev. 2018 Jan 22;1(1):CD011551. doi: 10.1002/14651858.CD011551.pub2. Cochrane Database Syst Rev. 2018. PMID: 29357120 Free PMC article.
-
Screening for aspiration risk associated with dysphagia in acute stroke.Cochrane Database Syst Rev. 2021 Oct 18;10(10):CD012679. doi: 10.1002/14651858.CD012679.pub2. Cochrane Database Syst Rev. 2021. PMID: 34661279 Free PMC article.
References
-
- Cintron M.A., Baumer Y., Pang A.P., Peterson E.M.A., Ortiz-Whittingham L.R., Jacobs J.A., Sharda S., Potharaju K.A., Baez A.S., Gutierrez-Huerta C.A., et al. Associations between the neural-hematopoietic-inflammatory axis and DNA methylation of stress-related genes in human leukocytes: Data from the Washington, D.C. cardiovascular health and needs assessment. Brain Behav. Immun. Health. 2025;45:100976. doi: 10.1016/j.bbih.2025.100976. - DOI - PMC - PubMed
-
- Nagao Y., Fujimoto M., Tian Y., Kameyama S., Hattori K., Hidese S., Kunugi H., Kanai Y., Arai E. Genome-wide DNA methylation profiling of blood samples from patients with major depressive disorder: Correlation with symptom heterogeneity. J. Psychiatry Neurosci. 2025;50:E112–E124. doi: 10.1503/jpn.240126. - DOI - PMC - PubMed
-
- Xia L., Luo X., Liang Y., Jiang X., Yang W., Yan J., Qi K., Li P. Epigenetic modifications of nuclear and mitochondrial DNA are associated with the disturbance of serum iron biomarkers among the metabolically unhealthy obesity school-age children. Nutr. J. 2025;24:51. doi: 10.1186/s12937-025-01108-6. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

