The Role of the RecFOR Complex in Genome Stability
- PMID: 40564904
- PMCID: PMC12192639
- DOI: 10.3390/ijms26125441
The Role of the RecFOR Complex in Genome Stability
Abstract
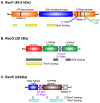
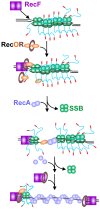
The maintenance of genome stability requires the coordinated actions of multiple proteins and protein complexes. One critical family of proteins is the recombination mediators. Their role is to facilitate the formation of recombinase nucleoprotein filaments on single-stranded DNA (ssDNA). Filament formation can take place on post-replicative ssDNA gaps as well as on 3'-tailed duplexes resulting from helicase-nuclease processing. In prokaryotes, the RecF, O, and R proteins are widely distributed and mediate RecA loading as either the RecFOR or RecOR complexes, depending on the species being studied. In this review, I compare and contrast the available biochemical and structural information to provide insight into the mechanism of action of this critical family of mediators.
Keywords: DNA repair; RecF; RecO; RecR; SSB protein; genetic recombination; recombination mediator.
Conflict of interest statement
The author declares no conflict of interest.
Figures



Similar articles
-
RecFOR and RecOR as distinct RecA loading pathways.J Biol Chem. 2009 Jan 30;284(5):3264-3272. doi: 10.1074/jbc.M807220200. Epub 2008 Nov 4. J Biol Chem. 2009. PMID: 18986990 Free PMC article.
-
Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein.J Biol Chem. 1994 Nov 25;269(47):30005-13. J Biol Chem. 1994. PMID: 7962001
-
The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins.Nucleic Acids Res. 2008 Jan;36(1):94-109. doi: 10.1093/nar/gkm1004. Epub 2007 Nov 13. Nucleic Acids Res. 2008. PMID: 18000001 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Education support services for improving school engagement and academic performance of children and adolescents with a chronic health condition.Cochrane Database Syst Rev. 2023 Feb 8;2(2):CD011538. doi: 10.1002/14651858.CD011538.pub2. Cochrane Database Syst Rev. 2023. PMID: 36752365 Free PMC article.
References
-
- Bianco P.R., Tracy R.B., Kowalczykowski S.C. DNA strand exchange proteins: A biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. - PubMed
-
- Bianco P.R. eLS. John Wiley & Sons, Ltd.; Hoboken, NJ, USA: 2001. RecA Protein; pp. 1–12.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

