Repurposing Biomolecules from Aerva javanica Against DDX3X in LAML: A Computer-Aided Therapeutic Approach
- PMID: 40564907
- PMCID: PMC12193238
- DOI: 10.3390/ijms26125445
Repurposing Biomolecules from Aerva javanica Against DDX3X in LAML: A Computer-Aided Therapeutic Approach
Abstract
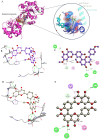
Acute Myeloid Leukemia (LAML) is a life-threatening hematological malignancy, and the DEAD-box helicase 3 X-linked (DDX3X) gene is a potential yet underexplored target gene for LAML. Biomolecules derived from medicinal plants like Aerva javanica offer a great source of therapeutic candidates. This study aimed to investigate the role of DDX3X in LAML and identify plant-derived biomolecules that could inhibit DDX3X using computational approaches. Pan-cancer mutational profiling, a transcriptomic analysis, survival, protein-protein interaction networks, and a principal component analysis (PCA) were employed to elucidate functional associations and transcriptomic divergence. Subsequently, biomolecules from A. javanica were subjected to in silico screening using molecular docking and ADMET profiling. The docking protocol was validated using RK-33, a known DDX3X inhibitor. DDX3X was found to be linked to key leukemogenic pathways, including Wnt/β-catenin and MAPK signaling, indicating it to be a potential target. Molecular docking of A. javanica compounds revealed CIDs 15559724, 5490003, and 74819331 as potent DDX3X inhibitors with strong binding affinity and favorable pharmacokinetic and toxicity profiles compared to RK-33. This study highlights the importance of DDX3X in LAML pathogenesis and suggests targeting it using plant-derived inhibitors, which may require further in vitro and in vivo validation.
Keywords: A. javanica; ADMET; DEAD-box helicase 3 X-linked (DDX3X); acute myeloid leukemia (LAML); molecular docking.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Exploring Type II Diabetes Inhibitors from Genus Daphne Plant-species: An Integrated Computational Study.Comb Chem High Throughput Screen. 2025;28(8):1413-1442. doi: 10.2174/0113862073262227231005074024. Comb Chem High Throughput Screen. 2025. PMID: 38584562
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
DEAD-box helicase intrinsically disordered domains and structural dynamics of HIV-1 RNA are required to reveal DDX3X catalytic efficiency.Nucleic Acids Res. 2025 Aug 27;53(16):gkaf834. doi: 10.1093/nar/gkaf834. Nucleic Acids Res. 2025. PMID: 40874589 Free PMC article.
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

