Proteomic Analysis of ARID1A-Deficient Ovarian Clear Cell Carcinoma Cells Reveals Differential Mitochondria ETC Subunit Abundances and Targetable Mitochondrial Pathways
- PMID: 40564930
- PMCID: PMC12193532
- DOI: 10.3390/ijms26125466
Proteomic Analysis of ARID1A-Deficient Ovarian Clear Cell Carcinoma Cells Reveals Differential Mitochondria ETC Subunit Abundances and Targetable Mitochondrial Pathways
Abstract
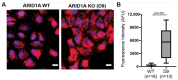
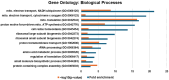
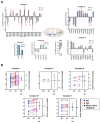
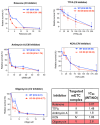
ARID1A-deficient ovarian clear cell carcinoma is a highly lethal gynecologic cancer that depends heavily on mitochondrial respiration. Our biochemical and proteomic analyses reveal that ARID1A knockout cells exhibit marked upregulation of specific subunits within mitochondrial electron transport chain (ETC) Complexes I, III, and IV. However, this upregulation does not directly translate into increased sensitivity to broad-spectrum inhibitors targeting these complexes. These findings suggest that broad-spectrum mitochondrial inhibitors may not be effective therapeutic options for ARID1A-deficient cancers. Instead, the selective inhibition of specific ETC subunits may offer a more promising approach to exploit the metabolic vulnerabilities of ARID1A-deficient cells.
Keywords: ARID1A; electron transport chain; mitochondria; ovarian clear cell carcinoma; proteomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Berns K., Caumanns J.J., Hijmans E.M., Gennissen A.M.C., Severson T.M., Evers B., Wisman G.B.A., Jan Meersma G., Lieftink C., Beijersbergen R.L., et al. ARID1A mutation sensitizes most ovarian clear cell carcinomas to BET inhibitors. Oncogene. 2018;37:4611–4625. doi: 10.1038/s41388-018-0300-6. - DOI - PMC - PubMed
-
- Katagiri A., Nakayama K., Rahman M.T., Rahman M., Katagiri H., Nakayama N., Ishikawa M., Ishibashi T., Iida K., Kobayashi H., et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod. Pathol. 2012;25:282–288. doi: 10.1038/modpathol.2011.161. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

