Multifaceted Roles of Guanylate-Binding Proteins in Cancer
- PMID: 40564939
- PMCID: PMC12193362
- DOI: 10.3390/ijms26125477
Multifaceted Roles of Guanylate-Binding Proteins in Cancer
Abstract
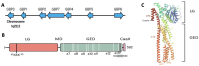
Guanylate-binding proteins (GBPs), encompassing GBP1 through GBP7 in humans, are interferon-inducible large GTPases of the dynamin superfamily, renowned for their pivotal roles in cell-autonomous immunity against intracellular pathogens such as viruses, bacteria, and protozoa. By recognizing pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs), GBPs orchestrate lysosomal targeting, regulate inflammatory cascades, and modulate apoptosis to protect host tissues from immune-mediated damage. Beyond their foundational roles in immunity, GBPs exhibit context-dependent effects in human cancer, promoting malignancy in some tumors through enhanced immune signaling, inhibition of apoptosis, and resistance to therapies, or suppressing tumor growth through immune activation and cell cycle regulation. This comprehensive review explores the structural intricacies, immune functions, and multifaceted contributions of human GBPs to cancer, delving into their molecular mechanisms, prognostic potential, and therapeutic implications. We incorporate the latest insights to highlight how understanding GBP regulation could reshape cancer treatment strategies.
Keywords: GTPase; cancer; guanylate-binding proteins; immunity.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Guanylate binding proteins (GBPs) as novel therapeutic targets against single-stranded RNA viruses.Mol Biol Rep. 2025 Aug 2;52(1):780. doi: 10.1007/s11033-025-10889-2. Mol Biol Rep. 2025. PMID: 40751764 Review.
-
Regulation of innate immune functions by guanylate-binding proteins.Int J Med Microbiol. 2018 Jan;308(1):237-245. doi: 10.1016/j.ijmm.2017.10.013. Epub 2017 Nov 2. Int J Med Microbiol. 2018. PMID: 29174633 Review.
-
Fungi and cancer: unveiling the complex role of fungal infections in tumor biology and therapeutic resistance.Front Cell Infect Microbiol. 2025 Jun 10;15:1596688. doi: 10.3389/fcimb.2025.1596688. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40557321 Free PMC article. Review.
-
Guanylate-binding proteins: emerging critical functions in plant development, immunity and stress response.New Phytol. 2025 Aug 29. doi: 10.1111/nph.70524. Online ahead of print. New Phytol. 2025. PMID: 40884028 Review.
-
Bacterial Extracellular Vesicles: Bridging Pathogen Biology and Therapeutic Innovation.Acta Biomater. 2025 Jun 15;200:1-20. doi: 10.1016/j.actbio.2025.05.028. Epub 2025 May 9. Acta Biomater. 2025. PMID: 40349898 Review.
References
-
- Britzen-Laurent N., Bauer M., Berton V., Fischer N., Syguda A., Reipschläger S., Naschberger E., Herrmann C., Stürzl M. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS ONE. 2010;5:e14246. doi: 10.1371/journal.pone.0014246. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

