Transglutaminase 2 Stimulates Cell Proliferation and Modulates Transforming Growth Factor-Beta Signaling Pathway Independently of Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells
- PMID: 40564959
- PMCID: PMC12192954
- DOI: 10.3390/ijms26125497
Transglutaminase 2 Stimulates Cell Proliferation and Modulates Transforming Growth Factor-Beta Signaling Pathway Independently of Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells
Abstract
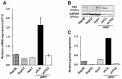
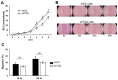
Transglutaminase 2 (TG2) is a multifunctional protein and plays a role in cancer progression. We previously identified TG2 as an early-recurrence biomarker in hepatocellular carcinoma (HCC). TG2-knockdown (shTG2) and control (shCtl) HCC cell lines were used for comparative analyses to clarify the molecular mechanisms underlying the contribution of this protein to HCC malignancy. The proliferation of shTG2 cells was slightly but significantly decreased compared with that of shCtl cells. Differential gene expression profiling based on GeneChip arrays revealed the enrichment of the PI3K-Akt signaling pathway and showed that the expression of Dickkopf-1 and -3 (DKK1 and DKK3, respectively), inhibitors and modulators of the Wnt/β-catenin signaling pathway, was increased in shTG2 cells. The expression of epithelial-mesenchymal transition (EMT)-related genes was similar in both shCtl and shTG2 cells before and after TGF-β1 treatment, even though TGF-β1 markedly upregulated TG2. Thus, TG2 may contribute to cancer malignancy via the stimulation of cell proliferation signaling, such as PI3K-Akt and Wnt/β-catenin signaling, but not EMT. This effect might be further enhanced by humoral factors such as TGF-β1 from the tumor microenvironment.
Keywords: PI3K-Akt signaling; TGF-β1; Wnt/β-catenin signaling; epithelial–mesenchymal transition; hepatocellular carcinoma; transglutaminase 2; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
TGM2-mediated histone serotonylation promotes HCC progression via MYC signalling pathway.J Hepatol. 2025 Jul;83(1):105-118. doi: 10.1016/j.jhep.2024.12.038. Epub 2025 Jan 7. J Hepatol. 2025. PMID: 39788430
-
CXCR2/CXCL5 axis contributes to epithelial-mesenchymal transition of HCC cells through activating PI3K/Akt/GSK-3β/Snail signaling.Cancer Lett. 2015 Mar 28;358(2):124-135. doi: 10.1016/j.canlet.2014.11.044. Epub 2014 Nov 24. Cancer Lett. 2015. PMID: 25462858
-
Aberrant expression of cuproptosis‑related gene LIPT1 is associated with metabolic dysregulation of fatty acid and prognosis in hepatocellular carcinoma.J Cancer Res Clin Oncol. 2023 Nov;149(17):15763-15779. doi: 10.1007/s00432-023-05325-6. Epub 2023 Sep 5. J Cancer Res Clin Oncol. 2023. PMID: 37668796 Free PMC article.
-
The Protective Role of miR-125b in Hepatocellular Carcinoma: Unraveling Tumor-Suppressive Mechanisms.Curr Mol Med. 2025;25(6):663-671. doi: 10.2174/0115665240304247240529074123. Curr Mol Med. 2025. PMID: 38859784 Review.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
References
-
- Teng W., Wu T.-C., Lin S.-M. Hepatocellular Carcinoma Systemic Treatment 2024 Update: From Early to Advanced Stage. Biomed. J. 2024;540:100815 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

