SARS-Cov-2 Replication in a Blood-Brain Barrier Model Established with Human Brain Microvascular Endothelial Cells Induces Permeability and Disables ACE2-Dependent Regulation of Bradykinin B1 Receptor
- PMID: 40565006
- PMCID: PMC12193337
- DOI: 10.3390/ijms26125540
SARS-Cov-2 Replication in a Blood-Brain Barrier Model Established with Human Brain Microvascular Endothelial Cells Induces Permeability and Disables ACE2-Dependent Regulation of Bradykinin B1 Receptor
Abstract
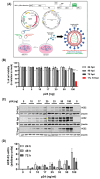
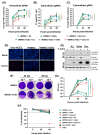
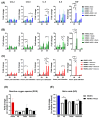
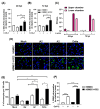
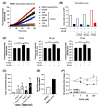
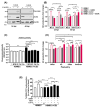
Endothelial dysfunction plays a central role in COVID-19 pathogenesis, by affecting vascular homeostasis and worsening thromboinflammation. This imbalance may contribute to blood-brain barrier (BBB) disruption, which has been reported in long COVID-19 patients with neurological sequelae. The kallikrein-kinin system (KKS) generates bradykinin (BK), a proinflammatory peptide that induces microvascular leakage via B2R. Under inflammatory conditions, BK is converted to Des-Arg-BK (DABK), which activates B1R, a receptor upregulated in inflamed tissues. DABK is degraded by ACE2, the main SARS-CoV-2 receptor; thus, viral binding and ACE2 downregulation may lead to DABK/B1R imbalance. Here, we investigated these interactions using human brain microvascular endothelial cells (HBMECs), as a model of the BBB. Since endothelial cell lines express low levels of ACE2, HBMECs were modified with an ACE2-carrying pseudovirus. SARS-CoV-2 replication was confirmed by RNA, protein expression, and infectious particles release. Infection upregulated cytokines and endothelial permeability, enhancing viral and leukocyte transmigration. Additionally, viral replication impaired ACE2 function in HBMECs, amplifying the response to DABK, increasing nitric oxide (NO) production, and further disrupting endothelial integrity. Our findings reveal a mechanism by which SARS-CoV-2 impacts the BBB and highlights the ACE2/KKS/B1R axis as a potential contributor to long COVID-19 neurological symptoms.
Keywords: ACE2; SARS-CoV-2; blood–brain barrier; des-Arg-bradykinin; endothelial cells; inflammation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;28:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- (01.20.0029.000462/20 404096/2020-4; 01.22.0074.00 (1227/21))/Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI
- CAPES/Coordination for the Improvement of Higher Education Personnel
- CNPq/Brazilian National Council for Scientific and Technological Development
- FAPERJ; 740 LBA E-26/201.206/2021, E-26/204.307/2024; JS E-26/ 210.059/ 2020, and E-26/210.251/-2020 and E-741 26/201.062/2021; SVAC E-26/20.746/2022; BBB E-26/202.346/2024; LRL E-26/201.918/2024/Carlos Chagas Filho Research Support Foundation
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

