Targeted DNA Methylation Using Modified DNA Probes: A Potential Therapeutic Tool for Depression and Stress-Related Disorders
- PMID: 40565107
- PMCID: PMC12193527
- DOI: 10.3390/ijms26125643
Targeted DNA Methylation Using Modified DNA Probes: A Potential Therapeutic Tool for Depression and Stress-Related Disorders
Abstract
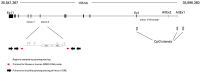
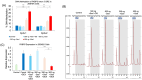
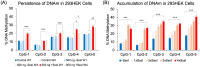
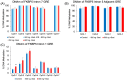
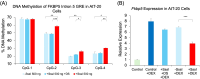
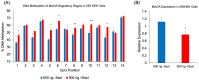
Epigenetic modifications play a crucial role in gene regulation and have been implicated in various physiological processes and disease conditions. DNA methylation (DNAm) has been implicated in the etiology and progression of many stress-related psychiatric behaviors, such as depression. The ability to manipulate DNAm may provide a means to reverse and treat such disorders. Although CRISPR-based technologies have enabled locus-specific DNAm editing, their clinical applicability may be limited due to immunogenicity concerns and off-target effects. In this study, we introduce a novel approach for targeted DNAm manipulation using single-stranded methylated DNA probes. The probes were designed against the GRE of FKBP5 and the promoter region of MAOA. In both human embryonic kidney HEK293 and mouse pituitary AtT-20 cells, transfection with their respective methylated probes significantly increased DNAm at targeted CpG sites in a persistent and dose-dependent manner. Importantly, the induced methylation effectively attenuated glucocorticoid-induced upregulation of FKBP5 gene expression. Alteration of methylation was specific to single-stranded probes, as double-stranded methylated probes and unmethylated probes showed no significant effects. Some limitations include the need to further characterize factors that influence probe efficiency, such as probe length and CpG density; develop an efficient in vivo probe delivery system; and perform a more extensive consideration of possible off-target effects. Despite these limitations, our findings suggest that methylated DNA probes have the potential to function as a simple tool for targeted epigenetic manipulation and serve as a safer alternative to CRISPR-based epigenome editing tools for the treatment of stress-related disorders such as depression.
Keywords: DNA methylation (DNAm); FK506 binding protein 5 (FKBP5); cortisol; epigenetics; gene expression; glucocorticoid response element (GRE); monoamine oxidase A (MAOA).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
[Epigenetics' implication in autism spectrum disorders: A review].Encephale. 2017 Aug;43(4):374-381. doi: 10.1016/j.encep.2016.07.007. Epub 2016 Sep 28. Encephale. 2017. PMID: 27692350 French.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Epigenetic editing and epi-drugs: a combination strategy to simultaneously target KDM4 as a novel anticancer approach.Clin Epigenetics. 2025 Jun 19;17(1):105. doi: 10.1186/s13148-025-01913-0. Clin Epigenetics. 2025. PMID: 40537846 Free PMC article.
-
Parent-training programmes for improving maternal psychosocial health.Cochrane Database Syst Rev. 2004;(1):CD002020. doi: 10.1002/14651858.CD002020.pub2. Cochrane Database Syst Rev. 2004. Update in: Cochrane Database Syst Rev. 2012 Jun 13;(6):CD002020. doi: 10.1002/14651858.CD002020.pub3. PMID: 14973981 Updated.
References
-
- Shireby G., Dempster E.L., Policicchio S., Smith R.G., Pishva E., Chioza B., Davies J.P., Burrage J., Lunnon K., Vellame D.S., et al. DNA methylation signatures of Alzheimer’s disease neuropathology in the cortex are primarily driven by variation in non-neuronal cell-types. Nat. Commun. 2022;13:1–14. doi: 10.1038/s41467-022-33394-7. - DOI - PMC - PubMed
-
- Yuan M., Yang B., Rothschild G., Mann J.J., Sanford L.D., Tang X., Huang C., Wang C., Zhang W. Epigenetic regulation in major depression and other stress-related disorders: Molecular mechanisms, clinical relevance and therapeutic potential. Signal Transduct. Target. Ther. 2023;8:309. doi: 10.1038/s41392-023-01519-z. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

