Tumor Microenvironment, Inflammation, and Inflammatory Prognostic Indices in Diffuse Large B-Cell Lymphomas: A Narrative Review
- PMID: 40565133
- PMCID: PMC12192974
- DOI: 10.3390/ijms26125670
Tumor Microenvironment, Inflammation, and Inflammatory Prognostic Indices in Diffuse Large B-Cell Lymphomas: A Narrative Review
Abstract
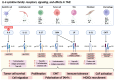
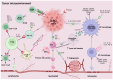
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of non-Hodgkin lymphoma, characterized by significant variability in clinical outcomes. Emerging evidence highlights the pivotal role of inflammation in the pathogenesis and prognosis of DLBCL. This narrative review explores the interplay between the tumor microenvironment, inflammatory processes, and prognostic indices used in DLBCL, focusing on biomarkers, immune responses, and systemic inflammation. These indices show promise as predictive and prognostic tools comparable to molecular markers, such as gene expression profiling, which are currently considered gold standards in prognosis but are often costly and technically demanding. By synthesizing findings from the current literature, this article highlights the potential of inflammatory indices as accessible and cost-effective prognostic alternatives to molecular markers in DLBCL, while also underscoring the need for further research to validate their clinical utility.
Keywords: DLBCL; adaptive immunity; diffuse large B-cell lymphoma; inflammation; innate immunity; prognostic index; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Campo E., Jaffe E.S., Cook J.R., Quintanilla-Martinez L., Swerdlow S.H., Anderson K.C., Brousset P., Cerroni L., de Leval L., Dirnhofer S., et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A report from the Clinical Advisory Committee. Blood. 2022;140:1229–1253. doi: 10.1182/blood.2022015851. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

