Two Peas in a Pod: Retroviral RNA Dimers Organize Gag-RNA Nanoclusters with Novel Biophysical Properties
- PMID: 40565141
- PMCID: PMC12193034
- DOI: 10.3390/ijms26125679
Two Peas in a Pod: Retroviral RNA Dimers Organize Gag-RNA Nanoclusters with Novel Biophysical Properties
Abstract
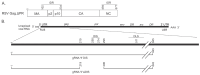
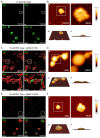
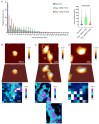
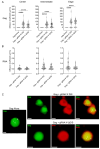
The continued effective control of retroviral infections will no doubt require the development of new clinical interventions targeting underexploited areas of retroviral biology such as genome selection and virion assembly. In our previous work, we demonstrated that both the Gag-psi (Ψ) interaction and genomic RNA (gRNA) dimerization each uniquely contribute to the formation, morphology, and stability of Rous sarcoma virus (RSV) Gag-viral RNA (vRNA) biomolecular condensates (BMCs). The present work builds upon those observations, utilizing atomic force microscopy (AFM) and fluorescence correlation spectroscopy (FCS) to elucidate the nanoscale morphology, resistance to mechanical deformation, and constituent diffusivity of RSV Gag-vRNA BMCs. These approaches revealed a novel role for gRNA dimerization in nanoscale condensate architecture and mechanical stability that aids in our understanding of why gRNA dimerization is critical for efficient packaging of the retroviral genome. Further biophysical characterization of RSV Gag-gRNA BMCs therefore possesses great potential to reveal novel avenues for therapeutic intervention.
Keywords: Rous sarcoma virus; atomic force microscopy; biomolecular condensates; biophysics; dimerization; fluorescence correlation spectroscopy; genome selection; retroviruses.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Update of
-
Two peas in a pod: retroviral RNA dimers organize Gag-RNA nanoclusters with novel biophysical properties.bioRxiv [Preprint]. 2025 Jun 11:2025.06.09.658653. doi: 10.1101/2025.06.09.658653. bioRxiv. 2025. Update in: Int J Mol Sci. 2025 Jun 13;26(12):5679. doi: 10.3390/ijms26125679. PMID: 40661432 Free PMC article. Updated. Preprint.
Similar articles
-
Two peas in a pod: retroviral RNA dimers organize Gag-RNA nanoclusters with novel biophysical properties.bioRxiv [Preprint]. 2025 Jun 11:2025.06.09.658653. doi: 10.1101/2025.06.09.658653. bioRxiv. 2025. Update in: Int J Mol Sci. 2025 Jun 13;26(12):5679. doi: 10.3390/ijms26125679. PMID: 40661432 Free PMC article. Updated. Preprint.
-
Role of the Psi Packaging Signal and Dimerization Initiation Sequence in the Organization of Rous Sarcoma Virus Gag-gRNA Co-Condensates.Viruses. 2025 Jan 13;17(1):97. doi: 10.3390/v17010097. Viruses. 2025. PMID: 39861886 Free PMC article.
-
Dynamic interactions of retroviral Gag condensates with nascent viral RNA at transcriptional burst sites: implications for genomic RNA packaging.bioRxiv [Preprint]. 2025 Jun 2:2025.01.11.632546. doi: 10.1101/2025.01.11.632546. bioRxiv. 2025. PMID: 39829876 Free PMC article. Preprint.
-
Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly.Semin Cell Dev Biol. 2019 Feb;86:129-139. doi: 10.1016/j.semcdb.2018.03.015. Epub 2018 Apr 1. Semin Cell Dev Biol. 2019. PMID: 29580971 Free PMC article. Review.
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

