Uropathogenic Escherichia coli Associated with Risk of Urosepsis-Genetic, Proteomic, and Metabolomic Studies
- PMID: 40565144
- PMCID: PMC12193174
- DOI: 10.3390/ijms26125681
Uropathogenic Escherichia coli Associated with Risk of Urosepsis-Genetic, Proteomic, and Metabolomic Studies
Abstract
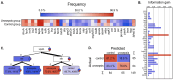
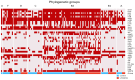
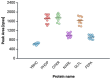
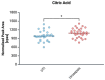
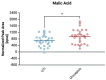
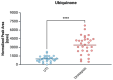
In the absence of fully effective therapies and preventive strategies against the development of urosepsis, a deeper understanding of the virulence mechanisms of Uropathogenic Escherichia coli (UPEC) strains is needed. UPEC strains employ a wide range of virulence factors (VFs) to persist in the urinary tract and bloodstream. UPEC strains were isolated from patients with sepsis and a control group without sepsis. PCR was used to detect 36 genes encoding various groups of virulence and fitness factors. Profiling of both intracellular and extracellular bacterial proteins was also included in our approach. Bacterial metabolites were identified and quantified using GC-MS and LC-MS techniques. The UpaG autotransporter, a trimeric E. coli AT adhesin, was significantly more prevalent in urosepsis strains (p = 0.00001). Iron uptake via aerobactin and the Iha protein also appeared to be predictive of urosepsis (p = 0.03 and p = 0.002, respectively). While some studies suggest an association between S fimbriae and the risk of urosepsis, we observed no such correlation (p = 0.0001). Proteomic and metabolomic analyses indicated that elevated levels of bacterial citrate, malate, coenzyme Q10, pectinesterase (YbhC), and glutamate transport proteins, as well as the regulators PhoP two-component system, CpxR two-component system, Nitrate/nitrite response regulator protein NarL, and the Ferrienterobactin receptor FepA, may play a role in sepsis. These genetic biomarkers, proteins, and metabolites derived from UPEC could potentially serve as indicators for assessing the risk of developing sepsis.
Keywords: Escherichia coli; OMICS technology; UPEC; markers; pathogenesis; sepsis; virulence.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

